Bioinformatics Programming
How to concatenate FASTA sequences using Perl?

Here is a simple Perl script to concatenate multiline FASTA sequences into a single line. A similar Perl script has been provided in one of the previous articles (A Perl script to convert multiline FASTA sequences into a single line). That script is useful for the FASTA files consisting of sequences split into a particular number of residues per line. The script provided in this article can also be used for large files consisting of multiple FASTA sequences split into the indefinite number of residues per line.
Let’s name the multiline FASTA input file as “input.fa”.
#!/usr/bin/perl -w
use strict;
my $input_fasta="input.fa";
open(FILE,"<$input_fasta") || die ("Error opening $input_fasta $!");
my $line = ;
print $line;
while ($line = )
{ chomp $line;
if ($line=~m/^>;(.*)/)
{ print "\n",$line,"\n"; }
else { print $line; }
}
print "\n";
exit;
Bioinformatics Programming
Free_Energy_Landscape-MD: Python package to create Free Energy Landscape using PCA from GROMACS.
In molecular dynamics (MD) simulations, a free energy landscape (FEL) serves as a crucial tool for understanding the behavior of molecules and biomolecules over time. It is difficult to understand and plot a meaningful FEL and then extract the time frames at which the plot shows minima. In this article, we introduce a new Python package (Free_Energy_Landscape-MD) to generate an FEL based on principal component analysis (PCA) from MD simulation done by GROMACS [1].
Bioinformatics News
VS_Analysis: A Python package to perform post-virtual screening analysis
Virtual screening (VS) is a crucial aspect of bioinformatics. As you may already know, there are various tools available for this purpose, including both paid and freely accessible options such as Autodock Vina. Conducting virtual screening with Autodock Vina requires less effort than analyzing its results. However, the analysis process can be challenging due to the large number of output files generated. To address this, we offer a comprehensive Python package designed to automate the analysis of virtual screening results.
Bioinformatics Programming
vs_interaction_analysis.py: Python script to perform post-virtual screening analysis
Analyzing the results of virtual screening (VS) performed with Autodock Vina [1] can be challenging when done manually. In earlier instances, we supplied two scripts, namely vs_analysis.py [2,3] and vs_analysis_compounds.py [4]. This time, we have developed a new Python script to simplify the analysis of VS results.
Bioinformatics Programming
How to create a pie chart using Python?
In this article. we are creating a pie chart of the docking score of five different compounds docked with the same protein. (more…)
Bioinformatics Programming
How to make swarm boxplot?
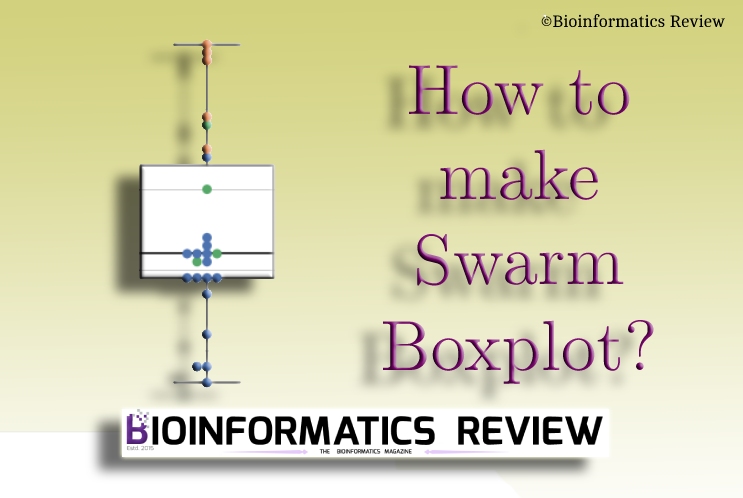
With the new year, we are going to start with a very simple yet complicated topic (for beginners) in bioinformatics. In this tutorial, we provide a simple code to plot swarm boxplot using matplotlib and seaborn. (more…)
Bioinformatics Programming
How to obtain ligand structures in PDB format from PDB ligand IDs?
Previously, we provided a similar script to download ligand SMILES from PDB ligand IDs. In this article, we are downloading PDB ligand structures from their corresponding IDs. (more…)
Bioinformatics Programming
How to obtain SMILES of ligands using PDB ligand IDs?
Fetching SMILE strings for a given number of SDF files of chemical compounds is not such a trivial task. We can quickly obtain them using RDKit or OpenBabel. But what if you don’t have SDF files of ligands in the first place? All you have is Ligand IDs from PDB. If they are a few then you can think of downloading SDF files manually but still, it seems time-consuming, especially when you have multiple compounds to work with. Therefore, we provide a Python script that will read all Ligand IDs and fetch their SDF files, and will finally convert them into SMILE strings. (more…)
Bioinformatics Programming
How to get secondary structure of multiple PDB files using DSSP in Python?
In this article, we will obtain the secondary structure of multiple PDB files present in a directory using DSSP [1]. You need to have DSSP installed on your system. (more…)
Bioinformatics Programming
vs_analysis_compound.py: Python script to search for binding affinities based on compound names.

Previously, we have provided the vs_analysis.py script to analyze virtual screening (VS) results obtained from Autodock Vina. In this article, we have provided another script to search for binding affinity associated with a compound. (more…)
Bioinformatics Programming
How to download files from an FTP server using Python?
In this article, we provide a simple Python script to download files from an FTP server using Python. (more…)
Bioinformatics Programming
How to convert the PDB file to PSF format?
VMD allows converting PDB to PSF format but sometimes it gives multiple errors. Therefore, in this article, we are going to convert PDB into PSF format using a different method. (more…)
Bioinformatics Programming
smitostr.py: Python script to convert SMILES to structures.
As mentioned in some of our previous articles, RDKit provides a wide range of functions. In this article, we are using RDKit [1] to draw a molecular structure using SMILES. (more…)
Bioinformatics Programming
How to preprocess data for clustering in MATLAB?
Data preprocessing is a foremost and essential step in clustering based on machine learning methods. It removes noise and provides better results. In this article, we are going to discuss the steps involved in data preprocessing using MATLAB [1]. (more…)
Bioinformatics Programming
How to calculate drug-likeness using RDKit?
RDKit [1] allows performing multiple functions on chemical compounds. One is the quantitative estimation of drug-likeness also known as QED properties. These properties include molecular weight (MW), octanol-water partition coefficient (ALOGP), number of hydrogen bond donors (HBD), number of hydrogen bond acceptors (HBA), polar surface area (PSA), number of rotatable bonds (ROTB), number of aromatic rings (AROM), structural alerts (ALERTS). (more…)
Bioinformatics Programming
sdftosmi.py: Convert multiple ligands/compounds in SDF format to SMILES.
You can obtain SMILES of multiple compounds or ligands in an SDF file in one go. Here, we provide a simple Python script to do that. (more…)
Bioinformatics Programming
tanimoto_similarities_one_vs_all.py – Python script to calculate Tanimoto Similarities of multiple compounds
We previously provided a Python script to calculate the Tanimoto similarities of multiple compounds against each other. In this article, we are providing another Python script to calculate the Tanimoto similarities of one compound with multiple compounds. (more…)
Bioinformatics Programming
tanimoto_similarities.py: A Python script to calculate Tanimoto similarities of multiple compounds using RDKit.
RDKit [1] is a very nice cheminformatics software. It allows us to perform a wide range of operations on chemical compounds/ ligands. We have provided a Python script to perform fingerprinting using Tanimoto similarity on multiple compounds using RDKit. (more…)
Bioinformatics Programming
How to commit changes to GitHub repository using vs code?

In this article, we are providing a few commands that are used to commit changes to GitHub repositories using VS code terminal.
Bioinformatics Programming
Extracting first and last residue from helix file in DSSP format.
Bioinformatics Programming
How to extract x,y,z coordinates of atoms from PDB file?

The x, y, and z coordinates of atoms are provided in the PDB file. One way to extract them is by using the Biopython package [1]. In this article, we will extract coordinates of C-alpha atoms for each residue from the PDB file using Biopython. (more…)









You must be logged in to post a comment Login