Etiology is the study of origination or causation of an event or phenomenon. Genetic etiology is the study of genes responsible for particular traits along with some other genes in an organism. The identification of genetic etiology has become a protocol while studying genotypes and/or phenotypes of individuals. For this, PRS which means, Polygenic Risk Score is calculated. A PRS is the summation of trait-associated alleles across various genetic loci, weighted by effect sizes on a trait of interest. The effect sizes are calculated by the Genome-Wide Association Studies (GWAS). It has revealed that the genetic basis of most of the complex traits caused by the small effects of hundreds or thousands of variants. The polygenic effects can be considered as the genetic liability to disease risk associated with these genes. PR score was found to be accurate in most of the applications. PRS only considers the variants with P-value threshold, i.e., PT.
PRS is calculated using estimated published GWAS results. This technique was first applied by the International Schizophrenia Consortium (2009), demonstrating that genetic risk for schizophrenia (SCZ) is a predictor of bipolar disorder. It has also been proved as the reliable indicator of genetic liability.
Jack Euesden et al. (2014), introduced a software package to easily calculate the PRS, known as PRSice (means ‘precise’).
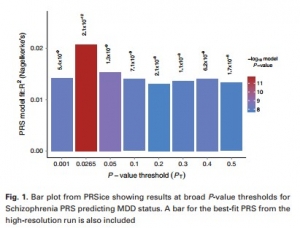
They found out the genetic relationship between the Schizophrenia (SCZ) and Major Depressive Disorder (MDD) and significantly proved that PRS of SCZ predicts the status of MDD.
PRSice returns the best fit PRS according to the polygenic risk associated with the alleles responsible for a trait. The main feature of PRSice is that it can easily and automatedly calculate the PRS at any value of P-value threshold, i.e., PT and then identifies the most precise threshold. It only requires the GWAS results of individuals on a base phenotype and genotype data on target phenotype, to calculate the PRS for each individual, and plots a PRS model depicting the fit range of PT value. PRSice can also consider the SNPs in linkage disequilibrium (i.e., an occurrence of a combination of genes in non-random proportions in a population), but it depends on the user whether to use this option or not. PRSice is a command line program that calculates PRS for individuals, under variously specified parameters and reduces the computation time.
For further reading, click here.
For any query, write to muniba@bioinformaticsreview.com
Reference:
PRSice: Polygenic Risk Score software —-Jack Euesden*, Cathryn M. Lewis, and Paul F. O’Reilly*