

RDP provides analysis tools called RDPTools. These tools are used to high-throughput sequencing data including single-strand, and paired-end reads [1]. In this article, we are going...

It is necessary to detect highly variable regions in envelopes of viruses as it allows the establishment of the viruses in the human body. A new...

Clustal packages [1,2] are quite useful in multiple sequence alignments. Especially, when you need specific outputs from the command-line. In this article, we will install CustalW2...
HMMER tool is used for searching sequence homologs using profile hidden Markov Models (HMMs) [1]. It is also one of the most widely used alignment tools....

FASTX-toolkit is a command-line bioinformatics software package for the preprocessing of short reads FASTQ/A files [1]. These files contain multiple short-read sequences obtained as an output...

DIAMOND is a program for high throughput pairwise alignment of DNA reads and protein sequences [1]. It is used for the high-performance analysis of large sequence...
MEME suite is used to discover novel motifs in unaligned nucleotide and protein sequences [1,2]. In this article, we will learn how to install MEME on...
BLAT is a pairwise sequence alignment algorithm that is used in the assembly and annotation of the human genome [1]. In this article, we will install...
NCBI-BLAST+ [1] command-line tool offers multiple functions to be performed on a large dataset of sequences. Previously, we have shown how to blast against a local...

Clustal Omega [1,2] and MUSCLE are bioinformatics tools that are used for multiple sequence alignment (MSA). In one of our previous articles, we explained the usage...
ClustalW2 is a bioinformatics tool for multiple sequence alignment of DNA or protein sequences. It can easily align sequences and generate a phylogenetic tree online (https://www.genome.jp/tools-bin/clustalw)....
PHMMER is a sequence analysis tool used for protein sequences (http://hmmer.org; version 3.1 b2). It is available online as a web server and as well as...

Sequence and structural data in bioinformatics are ever-increasing and the need for its analysis is ever-demanding likewise. As bioinformaticians analyze the data with their keen knowledge...


In our previous article, we discussed different multiple sequence alignment (MSA) benchmarks to compare and assess the available MSA programs. However, since the last decade, several...
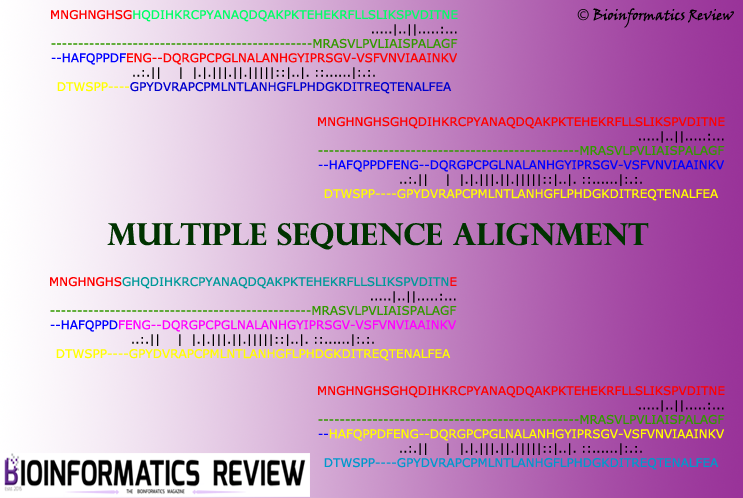

Multiple sequence alignment (MSA) is a very crucial step in most of the molecular analyses and evolutionary studies. Many MSA programs have been developed so far...

The basic local alignment search tool (BLAST) [1,2] is known for its speed and results, which is also a primary step in sequence analysis. The ever-increasing demand...

Sequence alignment is customary to not only find similar regions among a pair of sequences but also to study the structural, functional and evolutionary relationship between...
Latest Comments