Sequence Analysis
Multiple Sequence Alignment and Phylogenetic Tree construction using ClustalW2 command-line tool
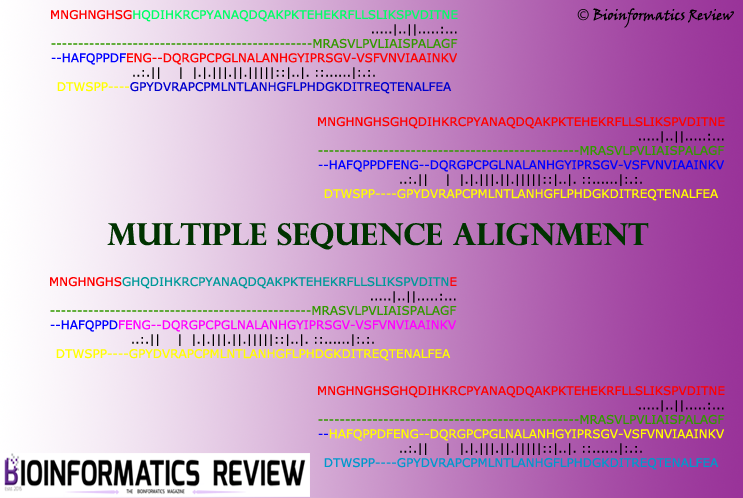
ClustalW2 is a bioinformatics tool for multiple sequence alignment of DNA or protein sequences. It can easily align sequences and generate a phylogenetic tree online (https://www.genome.jp/tools-bin/clustalw). However, in some cases, we need to perform these operations on a large number of FASTA sequences using the command-line tool of ClustalW2 [1]. It generates output files in very less time and provides quite accurate results. In this article, we will perform these operations using stand-alone tool of ClustalW2. Additionally, we will also generate a percent identity matrix (PIM) for the input sequences. PIM helps to identify the identity amongst the subjected sequences.
Let’s assume our input file name is ‘input.fasta’. We will run ClustalW2 on the Ubuntu platform in this article. If you wish to run on Windows, then enter the same command as mentioned below. Open the command prompt (cmd) on Windows and type the following command. Don’t forget to provide the full pathway of the ClustalW2 binary installed on your system.
Open a terminal (Ctrl+Alt+T) in Ubuntu and type the following commands:
$ /usr/local/bin/clustalw2 -infile=input.fasta -tree -pim -type=protein -case=upper
Provide full path to ClustalW2 binary, generally, it is /usr/local/bin/.
If you want your sequence residues to appear in small letters in the alignment, then type -case=lower and define the type of input sequences with -type argument.
It will generate .aln file as the alignment output, .tree as the phylogenetic tree output file, and .pim file as the PIM output.
References
- Higgins, D. G., & Sharp, P. M. (1988). CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene, 73(1), 237-244.
Sequence Analysis
HMMER- Uses & Applications

HMMER [1] is a well-known bioinformatics tool/software. It offers a web server and a command-line tool for users. Here are some additional applications of HMMER. (more…)
Sequence Analysis
Easy installation of some alignment software on Ubuntu (Linux) 18.04 & 20.04

There are commonly used alignment programs such as muscle, blast, clustalx, and so on, that can be easily installed from the repository. In this article, we are going to install such software on Ubuntu 18.04 & 20.04. (more…)
Sequence Analysis
FEGS- A New Feature Extraction Model for Protein Sequence Analysis
Protein sequence analyses include protein similarity, Protein function prediction, protein interactions, and so on. A new feature extraction model is developed for easy analysis of protein sequences. (more…)
Sequence Analysis
Installing RDPTools on Ubuntu (Linux)

RDP provides analysis tools called RDPTools. These tools are used to high-throughput sequencing data including single-strand, and paired-end reads [1]. In this article, we are going to install RDPTools on Ubuntu (Linux). (more…)
Sequence Analysis
NGlyAlign- A New Tool to Align Highly Variable Regions in HIV Sequences

It is necessary to detect highly variable regions in envelopes of viruses as it allows the establishment of the viruses in the human body. A new tool is developed to build and align the highly variable regions in HIV sequences. (more…)
Sequence Analysis
How to install ClustalW2 on Ubuntu?

Clustal packages [1,2] are quite useful in multiple sequence alignments. Especially, when you need specific outputs from the command-line. In this article, we will install CustalW2 command-line tool on Ubuntu. (more…)
Sequence Analysis
Installing HMMER package on Ubuntu
HMMER tool is used for searching sequence homologs using profile hidden Markov Models (HMMs) [1]. It is also one of the most widely used alignment tools. In this article, we will install the latest HMMER package on Ubuntu. (more…)
Sequence Analysis
Installing FASTX-toolkit on Ubuntu

FASTX-toolkit is a command-line bioinformatics software package for the preprocessing of short reads FASTQ/A files [1]. These files contain multiple short-read sequences obtained as an output of next-generation sequencing. In this article, we are going to install FASTX-toolkit on Ubuntu. (more…)
Sequence Analysis
Aligning DNA reads against a local database using DIAMOND

DIAMOND is a program for high throughput pairwise alignment of DNA reads and protein sequences [1]. It is used for the high-performance analysis of large sequence data. In this article, we will make a local database of protein sequences and align protein sequences against the reference database. (more…)
Sequence Analysis
Installing MEME suite on Ubuntu
MEME suite is used to discover novel motifs in unaligned nucleotide and protein sequences [1,2]. In this article, we will learn how to install MEME on Ubuntu. (more…)
Sequence Analysis
Installing BLAT- A Pairwise Alignment Tool on Ubuntu
BLAT is a pairwise sequence alignment algorithm that is used in the assembly and annotation of the human genome [1]. In this article, we will install BLAT on Ubuntu. (more…)
Sequence Analysis
Homology search against a local dataset using NCBI-BLAST+ command-line tool
NCBI-BLAST+ [1] command-line tool offers multiple functions to be performed on a large dataset of sequences. Previously, we have shown how to blast against a local dataset of sequences. This article will explain the search of homologous sequences for a query sequence against a local database of sequences and how to obtain the top 100 hits out of the searched results. (more…)
Sequence Analysis
How to use Clustal Omega and MUSCLE command-line tools for multiple sequence alignment?

Clustal Omega [1,2] and MUSCLE are bioinformatics tools that are used for multiple sequence alignment (MSA). In one of our previous articles, we explained the usage of the ClustalW2 command-line tool for MSA and phylogenetic tree construction. In this article, we will use Clustal Omega and MUSCLE for MSA exploring other arguments that facilitate different output formats. (more…)
Proteomics
Sequence search against a set of local sequences (local database) using phmmer
PHMMER is a sequence analysis tool used for protein sequences (http://hmmer.org; version 3.1 b2). It is available online as a web server and as well as a part of the HMMER stand-alone package (http://hmmer.org; version 3.1 b2). HMMER offers various useful features such as multiple sequence alignment including the file format conversion. (more…)
Sequence Analysis
Biotite: A bioinformatics framework for sequence and structure data analysis
Sequence and structural data in bioinformatics are ever-increasing and the need for its analysis is ever-demanding likewise. As bioinformaticians analyze the data with their keen knowledge and reach important conclusions, similarly, bioinformaticists provide with the enhanced and advanced tools and software for data analysis. (more…)
Algorithms
Simulated sequence alignment software: An alternative to MSA benchmarks

In our previous article, we discussed different multiple sequence alignment (MSA) benchmarks to compare and assess the available MSA programs. However, since the last decade, several sequence simulation software have been introduced and are gaining more interest. In this article, we will be discussing various sequence simulating software being used as alternatives to MSA benchmarks. (more…)
Algorithms
Benchmark databases for multiple sequence alignment: An overview
Multiple sequence alignment (MSA) is a very crucial step in most of the molecular analyses and evolutionary studies. Many MSA programs have been developed so far based on different approaches which attempt to provide optimal alignment with high accuracy. Basic algorithms employed to develop MSA programs include progressive algorithm [1], iterative-based [2], and consistency-based algorithm [3]. Some of the programs incorporate several other methods into the process of creating an optimal alignment such as M-COFFEE [4] and PCMA [5]. (more…)
Algorithms
SparkBLAST: Introduction

The basic local alignment search tool (BLAST) [1,2] is known for its speed and results, which is also a primary step in sequence analysis. The ever-increasing demand for processing huge amount of genomic data has led to the development of new scalable and highly efficient computational tools/algorithms. For example, MapReduce is the most widely accepted framework which supports design patterns representing general reusable solutions to some problems including biological assembly [3] and is highly efficient to handle large datasets running over hundreds to thousands of processing nodes [4]. But the implementation frameworks of MapReduce (such as Hadoop) limits its capability to process smaller data. (more…)
Algorithms
Role of Information Theory, Chaos Theory, and Linear Algebra and Statistics in the development of alignment-free sequence analysis

Sequence alignment is customary to not only find similar regions among a pair of sequences but also to study the structural, functional and evolutionary relationship between organisms. Many tools have been discovered to achieve the goal of alignment of a pair of sequences, separately for nucleotide sequence and amino acid sequence, BLOSSUM & PAM [1] are a few to name. (more…)
Sequence Analysis
A short introduction to protein structures modification and ModFinder

A lot of protein structures are determined on a large scale and submitted in Protein Data Bank (PDB) www.rcsb.org [1]. After the experimental determination of these structures, they are used in many scientific studies and experiments are performed upon them such as mutagenesis, docking, and so on. (more…)


![[Tutorial] Installing jdock on Ubuntu (Linux).](https://img.bioinformaticsreview.com/uploads/2023/05/09092952/jdock-400x240.jpg)
![[Tutorial] Installing jdock on Ubuntu (Linux).](https://img.bioinformaticsreview.com/uploads/2023/05/09092952/jdock-80x80.jpg)











You must be logged in to post a comment Login