wes_data_analysis: Whole Exome Sequencing (WES) Data visualization Toolkit
Whole Exome Sequencing (WES) is a genomic technique that sequences only the…
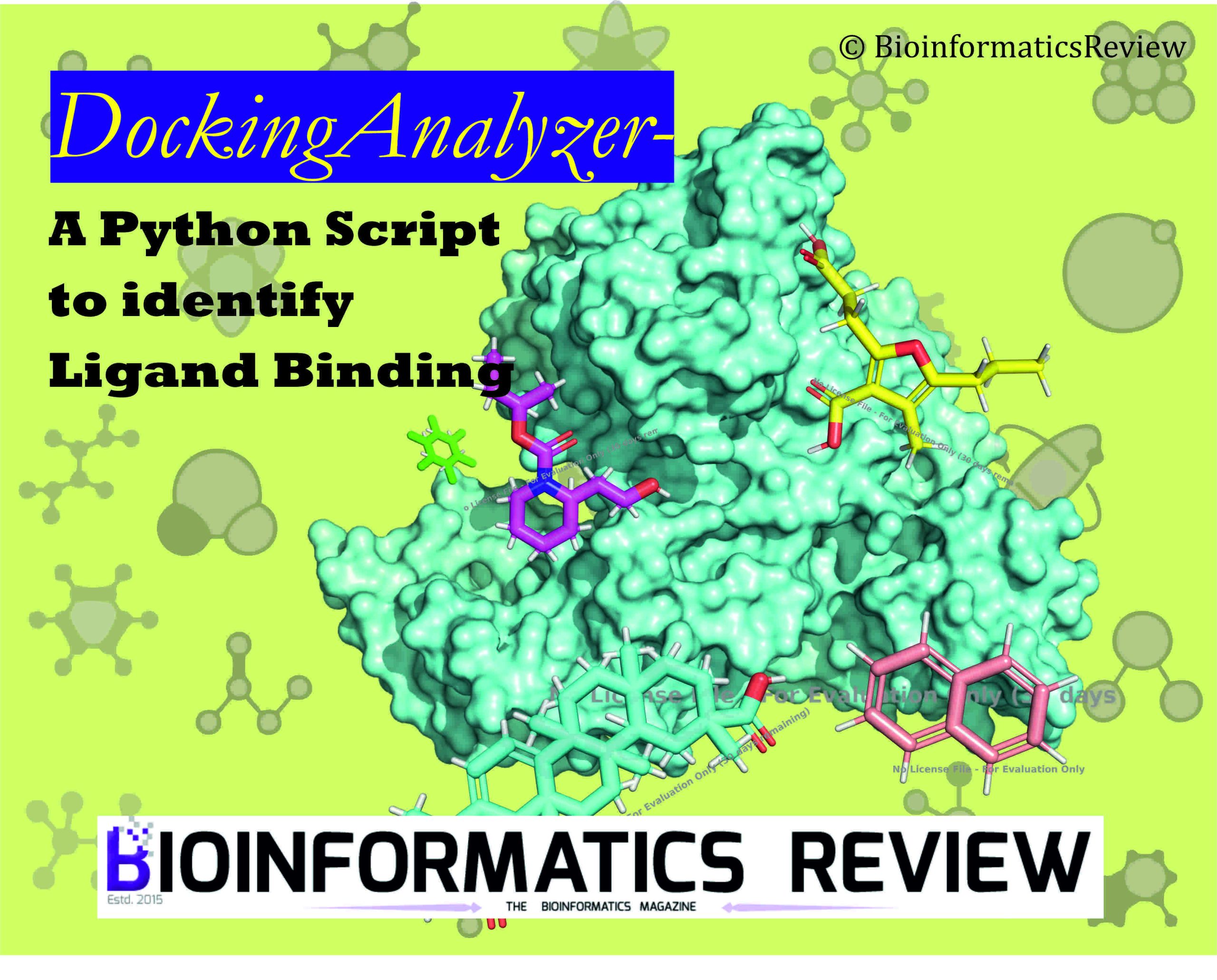
DockingAnalyzer.py: A New Python script to Identify Ligand Binding in Protein Pockets.
High-throughput virtual screening (HTVS) is a pivotal technique in drug discovery that…
Free_Energy_Landscape-MD: Python package to create Free Energy Landscape using PCA from GROMACS.
In molecular dynamics (MD) simulations, a free energy landscape (FEL) serves as…
VS_Analysis: A Python package to perform post-virtual screening analysis
Virtual screening (VS) is a crucial aspect of bioinformatics. As you may…
How to create a pie chart using Python?
In this article. we are creating a pie chart of the docking…
How to make swarm boxplot?
With the new year, we are going to start with a very…
How to obtain ligand structures in PDB format from PDB ligand IDs?
Previously, we provided a similar script to download ligand SMILES from PDB…
How to obtain SMILES of ligands using PDB ligand IDs?
Fetching SMILE strings for a given number of SDF files of chemical…
How to get secondary structure of multiple PDB files using DSSP in Python?
In this article, we will obtain the secondary structure of multiple PDB…
vs_analysis_compound.py: Python script to search for binding affinities based on compound names.
Previously, we have provided the vs_analysis.py script to analyze virtual screening (VS)…
How to download files from an FTP server using Python?
In this article, we provide a simple Python script to download files…
How to convert the PDB file to PSF format?
VMD allows converting PDB to PSF format but sometimes it gives multiple…
smitostr.py: Python script to convert SMILES to structures.
As mentioned in some of our previous articles, RDKit provides a wide…
How to calculate drug-likeness using RDKit?
RDKit allows performing multiple functions on chemical compounds. One is the quantitative…
sdftosmi.py: Convert multiple ligands/compounds in SDF format to SMILES.
You can obtain SMILES of multiple compounds or ligands in an SDF…
tanimoto_similarities_one_vs_all.py – Python script to calculate Tanimoto Similarities of multiple compounds
We previously provided a Python script to calculate the Tanimoto similarities of…
tanimoto_similarities.py: A Python script to calculate Tanimoto similarities of multiple compounds using RDKit.
RDKit is a very nice cheminformatics software. It allows us to perform…
Extracting first and last residue from helix file in DSSP format.
Previously, we have provided a tutorial on using dssp_parser to extract all…
How to extract x,y,z coordinates of atoms from PDB file?
The x, y, and z coordinates of atoms are provided in the…
dssp_parser: A new Python package to extract helices from DSSP files.
A new Python package named 'dssp_parser' is developed to parse DSSP files.…