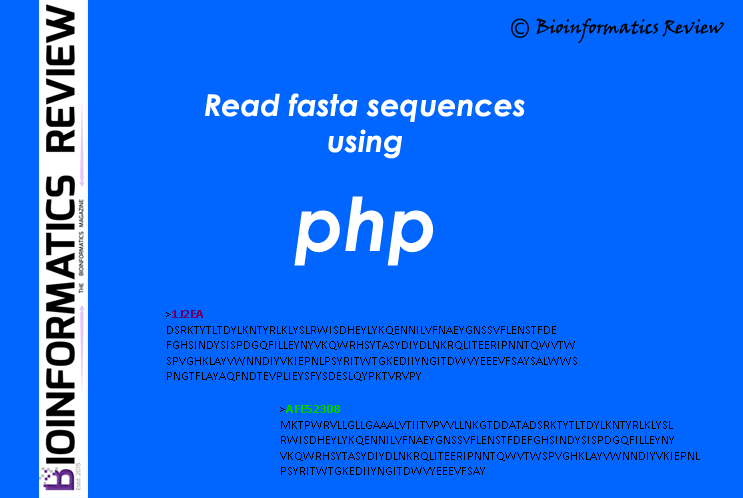
Here is a simple function in PHP to read fasta sequences from a file.
Your multifasta input file is “input.fasta”.
function read_fastas($filename){
$fh = fopen($filename, 'r');
$i= 0 ;
$sequences = array();
while($line = fgets($fh)){
$i++;
if($i%2==1){
$sequence['header'] = $line;
}
else{
$sequence['sequence'] = $line;
array_push($sequences, $sequence);
}
}
return $sequences;
}