CRISPR/Cas9 system is a bacterial defence mechanism against bacteriophage infection. When a viral dna(Bacteriophage, in this case) integrates into the bacterial genome, it produces RNA which is taken up by Cas9. Cas9 and the RNA together floats and drifts through the cell and as soon as they encounter a sequence complementary to the RNA, it gets attached to it. Cas9 chops off the dna from there. As the viral DNA is chopped off, it prevents the virus from multiplying. Thus the bacteria defends itself by precisely snipping out the viral DNA from its genome using CRISPR/Cas9 system.
The recent implementation of CRISPR/Cas9 system in human beings, animals and bacteria for gene editing has led to a lot of interesting research in this area. It requires designing of sgRNA known as Single Guide RNA, which is a challenging process. To solve this problem, CRISPR-ERA was developed.
So what is CRISPR-ERA?
CRISPR-ERA is a new tool available at http://crispr-era.stanford.edu developed by Honglei Liu et al. It is an acronym for Clustered Regularly Interspaced Short Palindromic Repeat-mediated Editing, Repression, and Activation.
What does CRISPR-ERA do?
According to the author of CRISPR-ERA,
The major goal of our designer tool is to address the discrepancy for designing sgRNAs that allow efficient and highly specific repression or activation of genes and for generating genome-wide sgRNA libraries for genetic screening in different organisms.
– Bioinformatics, 31(22), 2015, 3676–3678 doi: 10.1093/bioinformatics/btv423 (Paper)
How does CRISPR-ERA work?
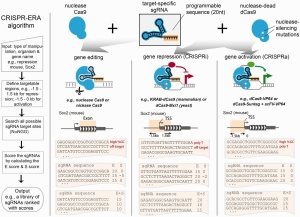 CRISPR-ERA looks up all targetable sites for each target gene, for patterns of N20NGG (N = any nucleotide). It then calculates E and S score.
CRISPR-ERA looks up all targetable sites for each target gene, for patterns of N20NGG (N = any nucleotide). It then calculates E and S score.
- E-score is the efficacy score ]based on the sequence features such as GC content (%GC), presence of poly-thymidine and location information
- S-score is the specificity score based on the genome-wide off-target binding sites. For each sgRNA design, enome-wide sequences are computed that contain an adjacent NRG (R = A or G) protospacer adjacent motif (PAM) site and zero, one, two, or three mismatches complementary to the sgRNA using Bowtie, which are regarded as off-target binding sites. The penalty score for NAG off-target is smaller than NGG off-target. The sgRNAs are finally ranked by the sum of E-score and S-score
The result it then presented according to the E and S score.
References & Further Reading
- http://gizmodo.com/everything-you-need-to-know-about-crispr-the-new-tool-1702114381
- Bioinformatics (2015) 31 (22):3676-3678.doi:10.1093/bioinformatics/btv423




Does Cas9 already present in our body or it is introduced externally?
Hi Muniba, Thank you for your interest in knowing more about Cas9 protein.
Cas9 is found naturally in Bacterial cells. It has evolved as a bacterial defence mechanism against bacteriophage.
ok thank you.