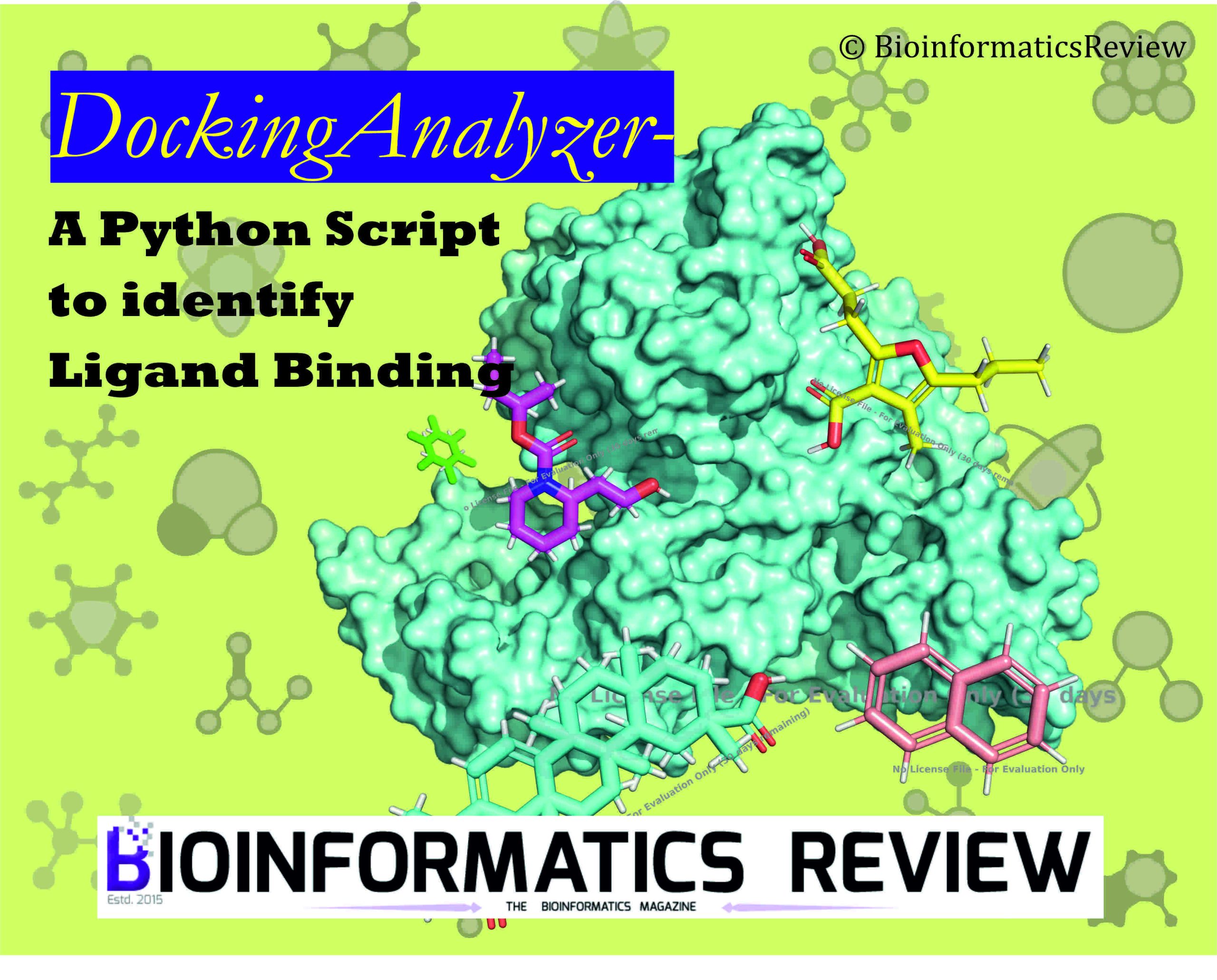
High-throughput virtual screening (HTVS) is a pivotal technique in drug discovery that screens extensive libraries of compounds to find potential drug candidates. One of the essential tasks in HTVS is to ensure that ligands are binding within the protein’s binding pocket. This task can be particularly challenging when dealing with thousands of docking results. To address this challenge, we present a Python script that automates the analysis of molecular docking results generated by AutoDock Vina [1] using PyMOL [2]. This script calculates the center of mass (COM) for each docked pose, compares it with a reference ligand’s COM, and identifies poses that bind within a specified threshold distance. This process is crucial in mass docking scenarios where confirming ligand binding within the pocket is necessary.
- Script Functions
- References
- How to Cite:
- Faiza, M., (2024). dockinganalyzer-py: a-new-python-script-to-identify-ligand-binding-in-protein-pockets, 10(5): page 12-16. https://bioinformaticsreview.com/20240110/vs_analysis-a-python-package-to-perform-post-virtual-screening-analysis/
- Faiza, M., (2024). VS_Analysis: A Python package to perform post-virtual screening analysis, 10(1): page 8-12. https://bioinformaticsreview.com/20240110/vs_analysis-a-python-package-to-perform-post-virtual-screening-analysis/
Background & Introduction
High-throughput virtual Screening (HTVS) has become a cornerstone in modern drug discovery, enabling researchers to evaluate vast libraries of chemical compounds efficiently. The goal of HTVS is to identify potential drug candidates that can interact effectively with a biological target, typically a protein associated with a disease. This process involves computationally simulating the interaction between each compound (ligand) and the target protein to predict their binding affinity and interaction modes.
In the realm of computational drug discovery, high-throughput virtual screening (HTVS) is a critical technique for identifying potential drug candidates from vast compound libraries. One of the most widely used tools for molecular docking in HTVS is AutoDock Vina [1]. AutoDock Vina is open-source and known for its speed and accuracy in predicting ligand binding conformations and affinities. Despite its effectiveness, analyzing the massive output from HTVS poses a significant challenge. However, one of the major challenges is to efficiently and accurately filter through the docked compounds to identify those that are likely to bind effectively within the target protein pocket. Specifically, during the filtering of docked compounds, we often consider only the binding affinity or docking score. However, each ligand will give a binding affinity/docking score irrespective of its binding within the pocket. This would give unnecessary compounds during the filtering process. To identify an effective compound/drug candidate, it is important to know if it is binding within the pocket or not. This script, named “DockingAnalyzer“, addresses this issue by calculating the center of mass (COM) of each docked ligand and comparing it to the COM of a reference ligand to determine if the ligand is truly binding within the target pocket.
DockingAnalyzer addresses this challenge by automating the post-processing of docking results. It leverages the powerful visualization and computational capabilities of PyMOL [2], a popular molecular visualization system, to calculate the center of mass (COM) for each docked pose and compare it to a reference ligand’s COM. This comparison helps identify which ligands are likely binding within the desired pocket, facilitating the selection of promising drug candidates for further experimental validation.
Script Overview
DockingAnalyzer is designed to process large numbers of docking output files generated by molecular docking software such as AutoDock Vina. The script calculates the center of mass (COM) for each ligand pose and compares it to the COM of a reference ligand. If the distance between the COM of the ligand pose and the reference ligand is below a specified threshold, the ligand pose is considered to be near the reference ligand and flagged accordingly. The script performs the following tasks:
- Loading Structures: The script begins by loading the receptor structure and the reference ligand into PyMOL. It calculates the reference ligand’s center of mass (COM) and uses it as a benchmark for comparison.
- Batch Processing: If the number of docking output files (in
.pdbqtformat) exceeds 100, the script processes them in batches. This ensures efficient memory management and prevents system overloads during large-scale screenings. - Calculating COMs: For each docking output file, the script splits the file into individual poses and calculates the COM for each pose. The COMs are rounded to three decimal places for consistency and easier comparison.
- Comparing COMs: The script calculates the Euclidean distance between the COM of each pose and the reference ligand’s COM. This distance (or difference magnitude) helps determine how close each pose is to the reference ligand’s binding position.
- Threshold-Based Filtering: A threshold distance (default is 4.0 Angstroms) is used to identify poses that are sufficiently close to the reference ligand’s COM. Poses with a distance less than the threshold are considered “near” the reference ligand, indicating they likely bind within the pocket. Users can adjust this threshold based on their requirements.
- Output Generation: The script generates a tab-separated values (TSV) file containing detailed information for each pose, including the ligand name, pose name, COM, difference magnitude, and a flag indicating whether the pose is near the reference ligand. Additionally, poses that are near the reference ligand are also written in a separate file for easy identification and further analysis.
Script Functions
The script contains two helper functions (calculate_COM and process_files) and a main function.
- calculate_COM: This function calculates the COM for each pose, compares it with the reference ligand COM, and writes the results to output files.
- process_files: This function processes the docking files in batches to manage large datasets efficiently. It loads the Vina output files one by one, splits their poses, and calculates the COM of each pose. It doesn’t mean that you can’t run it for a small number of docked compounds; it will work effectively for smaller datasets as well.
- main(): The
mainfunction orchestrates the entire process.
How DockingAnalyzer Works?
- Initialization: The script begins by accepting command-line arguments for the receptor and reference ligand files. It calculates the COM for the reference ligand using PyMOL’s computational tools. PyMOL is a molecular visualization system that allows for interactive exploration of molecular structures.
- File Handling: The script scans the current directory for all
.pdbqtfiles, which are the output format of AutoDock Vina. It counts these files and determines if they need to be processed in batches to handle large datasets efficiently. This batching mechanism ensures that the script can process thousands of files without overwhelming system resources. - Processing Docking Results: For each batch of docking files, the script:
- Loads the receptor and reference ligand into PyMOL.
- Splits the docking results into individual poses.
- Calculates the COM for each pose.
- Compares the pose COM with the reference ligand COM and determines if they are within a specified threshold.
- Writes the results to an output file, indicating if the pose is near the reference ligand.
- Output Generation: Results are written to two files:
hts-docking.txtcontains all processed poses with their COM and distance from the reference ligand COM.near_ref_ligand.txtcontains only those poses that are near the reference ligand. This file is crucial for quickly identifying ligands that are likely to bind effectively within the target pocket.
Availability
This Python script is a part of the VS-Analysis package. It is freely accessible on GitHub.
Usage
Keep all Vina output (.pdbqt) files in a directory including the PDB file of your receptor protein and PyMOL script (center_of_mass.py, provided with the package) [2]. Run the script using the following command:
python3 DockingAnalyzer.py <receptor_filename> <reference_ligand_filename>
- receptor_filename is the name of the receptor file in PDB format.
- reference_ligand_filename is the name of the Vina output reference ligand file in PDBQT format.
For example,
python3 DockingAnalyzer.py protein.pdb ref_ligand.pdbqt
Key Benefits
- Efficiency: Automates the processing of large numbers of docking output files, making the analysis much faster.
- Accuracy: Uses COM calculations and a threshold to ensure only ligands binding near the reference ligand are considered, reducing false positives.
- Scalability: Handles large datasets by processing files in batches, preventing memory overload and ensuring smooth execution.
- Practicality: Outputs detailed results in an easily readable TSV format, facilitating further analysis and decision-making.
Applications
This script is highly beneficial for researchers in the field of computational chemistry and drug discovery. Here are some potential applications:
- Drug Discovery: By automating the analysis of molecular docking results, researchers can quickly identify promising drug candidates.
- Structure-Based Drug Design: The script helps understand the binding interactions between ligands and the target protein, which is crucial for designing effective drugs.
- High-Throughput Screening: The batch processing capability efficiently handles large datasets, making it suitable for HTVS campaigns.
- Binding Affinity Studies: Researchers can use the script to study the binding affinities of different ligands and identify the most potent ones.
- Ensuring Binding within Pocket: The script’s ability to confirm if a ligand is binding within the protein’s active pocket is crucial in mass docking experiments, where many ligands are screened, and not all are guaranteed to bind within the desired site.
Conclusion
The presented script provides a robust and efficient way to automate the analysis of molecular docking results using PyMOL and Python. By calculating the COM of docked poses and comparing them with a reference ligand, the script helps identify potential drug candidates structurally similar to known active compounds. This automation saves time and enhances the accuracy of HTVS campaigns, making it an invaluable tool in the drug discovery pipeline. Moreover, its ability to ensure that ligands are binding within the protein’s active pocket makes it particularly useful in large-scale docking projects where manual verification would be impractical.
References
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry, 31(2), 455-461.
- The PyMOL Molecular Graphics System, Version 2.5, Schrödinger, LLC.