
So, today is the great festival of Christmas……! Birthday of The Son of God.. And on this Auspicious day, We want to present before you all...
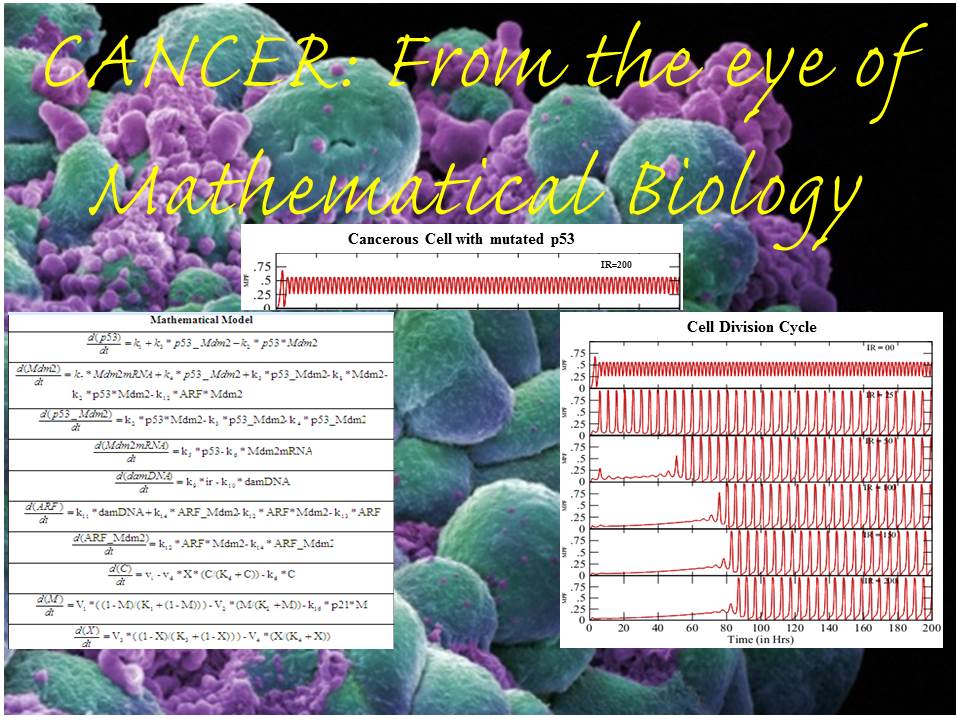

The month of November has just arrived with its generic glimpse of winter. We welcome this month with an evergreen and hot topic of cancer research. This time...
Latest Comments