Active learning is a kind of machine learning. Basically in active learning, a learning algorithm is used to perform the desired experiments to produce a desired output.
Active learning is a powerful tool for drug discovery and development where it reduces the tedious process of performing a number of experiments which are required to produce significant high-confidence predictions. However, practically it is difficult to decide when to stop the experimentation process. Therefore, if a reliable stopping criteria is applied to the algorithm reduces both time and cost of the experimentation process.
The basic of active learning is having good predictive models to guide experimentation.
Active learning iteratively builds a model for drug-target interactions. Instead of relying on large training data sets as performed manually, the active learning procedure increases the training set step wise. Thus, the time and experimental cost is reduced and it is only spent on improving the model rather than for the verification of a specific model which even may not be the desired outcome or suits the specifications under consideration.
How active learning works?
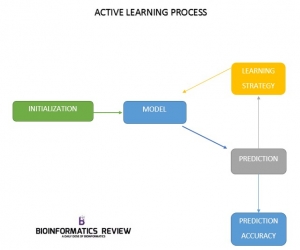
Active learning is an iterative process and is completed in four steps:
- Initialization
- Model
- Active learning algorithm
- Accuracy measure of the predicted output
The active learning strategy starts with an initialization step in which an interaction matrix for drug and target is formed. With the help of this matrix subset of known labels for the the drug and target kernels Kd and Kt respectively are provided. The model predicts the drug-target interactions. Based on the obtained predictions, the active learning algorithm is applied to find new experiments (labels) which will improve the model according to the requirements. Here, batchwise learning is applied where a fixed number of experiments is queried in each training round and thereby increases the size of experiments (labels). Each training round has a specific time point and is measured by the number of experiments performed. For each time point the accuracy of the model is predicted by using various methods. The process is stopped on some conditions, for example, if a certain budget for performing experiments is reached or the predicted accuracy of the model is high enough.
This is the basic idea for active learning applied in drug-target predictions. It saves a lot of time and cost involved in performing experiments in vitro. For further reading click here
Note:
An exhaustive list of references for this article is available with the author and is available on personal request, for more details write to muniba@bioinformaticsreview.com.









Great article Muniba, It is a really nice reading. It would be nice if you shed some light on the algorithm itself.Thank you!
Thanks for appreciation Tariq and I will explain the algorithm along with some stopping rules involved in drug-target interaction prediction in my upcoming article.
Thank You!