It all begins with finding a topic to work on! Find yourself a topic which has current relevance and with publications of clinical trials, case-control or response-no response studies in recent years, Randomized control trials are the gold standard for such analyses and are also hard to find. The procedure emphasizes the studies being recent as that makes the data for analysis updated to the latest methods, techniques, and error minimization procedures. Also, the recentness of included studies empowers the analyst’s view of the relevance of the topic in current times.
To construct a systematic review and conduct a meta-analysis of all literature published on any topic, one must know how to precisely extract meta-data from varied databases at their disposal. One may create his corpus of experimental studies from any single database, say PubMed, by submitting a query containing words/phrases outlining names of a disorder, prescribed/ on trial drugs, and type of study expected all connected by use of operators. PubMed usually will return with results in five-figures and this is a fairly large dataset to sort out manually.
We must consider at this point that PubMed is not the ultimate collection of all the biological literature ever published and in current publications, it is a sufficiently well-informed database but mustn’t be regarded as complete. There are other databases which act as topic-specific databases and can be expected to contain a more concentrated literature on the theme they nurture.
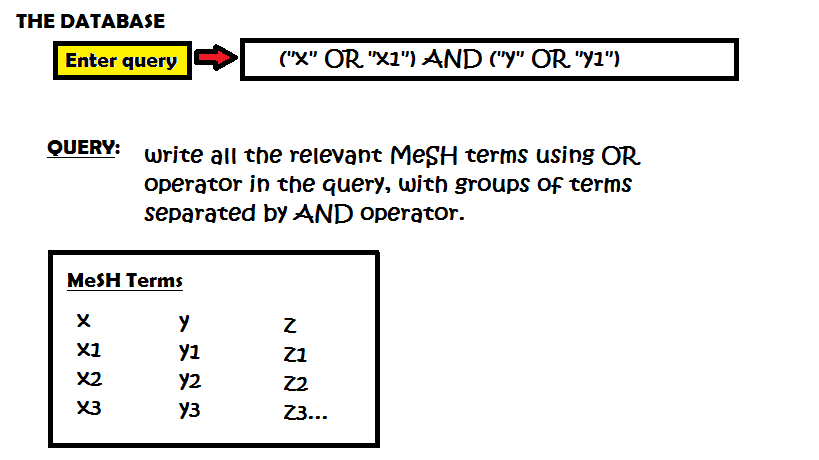
Moving on to the actual development of query, the analyst must ascertain that the search string developed is complete in all respects, i.e. it contains all the MeSH terms for the items queried. Even though PubMed will automatically generate synonyms for a query from its database of MeSH terms in our experience we found providing the MeSH terms saw a steep fall in the number of results returned, the drop was so significant that previously obtained five-figured corpus was reduced to a measly three figured and was manually sortable.
It is noteworthy that if possible one mustn’t filter out studies published in languages other than English as it may make the study biased by eliminating participation of regional studies and data. The analyst may always try to contact the authors for the English version of the study and thus reduce the amount of bias, making the analysis robust.
NOTE:
For any query write to manish@bioinformaticsreview.com