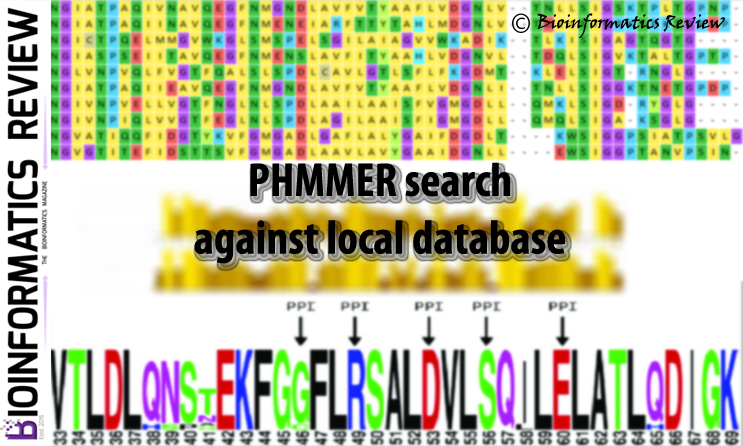
PHMMER is a sequence analysis tool used for protein sequences (http://hmmer.org; version 3.1 b2). It is available online as a web server and as well as a part of the HMMER stand-alone package (http://hmmer.org; version 3.1 b2). HMMER offers various useful features such as multiple sequence alignment including the file format conversion.
In this article, a sequence search against a set of local sequences is explained using PHMMER stand-alone tool including the output in FASTA format. To do this, we will first obtain the primary output in Stockholm (.sto) format and then convert it into the FASTA format.
1. Make a local database
The local database consists of protein sequences in FASTA format. Let’s say, our local dataset file is ‘sequences.fasta’.
2. Search for protein sequences according to the input in the local database
Make a query sequence file, we will name it as ‘query.fasta’. This file consists of FASTA sequences to be searched within the local database. Open a terminal and type the following command:
$ /path/to/phmmer -A phmmer.sto query.fasta sequences.fasta
where -A is used to define a filename to save the multiple alignments of all significant hits in Stockholm format.
You can also adjust the inclusion thresholds of different e-values by using different arguments. For example,
–incE, default value is 0.01 which means that ~1 false positive in every 100 searches with different query sequences.
–incT, instead of using e-value, use a bit score of >=<value>.
There are several other arguments that you can find in the user guide of HMMER.
Now, we have output in Stockholm format. If you want it in FASTA format, then proceed to the next step.
3. Output in FASTA format
For this, we will be using the ‘esl-reformat’ binary of HMMER
$ /path/to/esl-reformat fasta phmmer.sto > phmmerout.fasta
here, you can convert it into other formats such as a2m, just replace ‘fasta’ with ‘a2m’ in the command line.
This output file will consist of FASTA sequences of significant hits.