FOXL2 gene codes for a protein belonging to the fork-head- winged helix transcription factor. The FOXL2 gene is present on chromosome 3 and is a single exon gene of 2.7 kb coding for a protein comprising of 376 amino acids [1]. The protein has a DNA-binding domain, 110 amino acids long with a polyalanine tract. This polyalanine tract is restrained strictly at 14 residues. The reason for this conservation may be attributed to the optimal transcription factor activity occurring at that length. In addition, it has been observed that the FOXL2 protein is highly conserved across human, goat, mouse, and pufferfish [2]. However, it has been observed that the C-terminal end is more conserved as compared to the N-terminal end [3].
Structure of the FOXL2 protein
The FOXL2 protein has a unique DNA binding domain which is called as forkhead (FHD) [1]. This FHD is conserved among the other FOX proteins. It has a helix-turn-helix motif with 3 α-helices (H1, H2, and H3) and two large loops or “wings” (W1 and W2) [4]. These are the general components of the FHD of all FOX proteins. In the case of FoxL2 protein, there is an additional α-helix, H4, between H2 and H3.
Location of the FOXL2 protein
The FOXL2 protein is found in the developing eyelids and in fetal and adult ovaries of human, goat, and mouse. It is localized in the nucleus which can be attributed to the fact that it is a transcription factor. In the case of eyelids, it is present in the primordial mesenchyma. Additionally, unique to mice, the FOXL2 gene is also expressed in the pituitary Rathke’s pouch [5] and is said to be playing a role in the organogenesis of the pituitary gland.
In the ovaries, FOXL2 is expressed even before folliculogenesis and is present until the adult stages. It is localized in the somatic region with strong expression being observed in the follicular cells and a diffused expression being observed in the stromal cells [6]. During early ovarian development, FOXL2 is responsible for the determination/differentiation of the somatic cells and in later stages it is involved in the maintenance of ovarian function.
Consequences of FOXL2 mutations
Mutations in the FOXL2 gene are shown to cause Blepharophimosis Ptosis Epicanthus Inversus Syndrome (BPES). There are two types of BPES – Type 1 is characterized by eyelid malformation premature ovarian failure and Type 2 is characterized by only premature ovarian failure (POF) [7]. An increase (30% of mutations) in the number of residues in the polyalanine tract leads to BPES [8]. The mutation of the gene in BPES causes atrophy or hypertrophy of the superior levator muscles [9] and other periocular muscles which in turn lead to strabismus, a common observation among patients with BPES [10]. These arise from the primordial mesenchyma and the surrounding regions [11].
Sexual dimorphic expression of FOXL2
In females, Y chromosome (which contains the SRY gene) is absent and in such a condition FOXL2 is expressed leading to the development of granulosa cells [12]. In chicken, FOXL2 is expressed in ZW female gonads during days 5 to 8 whereas it is not the case in males during the same stages. In the case of turtles, where the sex of the organism is determined by the environmental temperature, the FOXL2 expression is higher in female promoting temperatures than in male promoting temperatures [13]. Therefore, clear sexual dimorphism can be noted in the expression of the FOXL2 gene.
FOXL2 targets and interactions
FOXL2 is involved in various cellular pathways like apoptosis, inflammation, antioxidation, etc., and also in the production of progesterone and estrogen, the significance of which will be discussed in detail in further sections.
Functions of FOXL2
Role in cholesterol metabolism
It is involved in the expression of Gonadotropin-releasing hormone (GnRH) receptor. The GnRH receptor activating sequence (GRAS) has binding sites for 3 transcriptional factors namely FOXL2, Smad3, and AP1 [14]. Additionally, it has been shown that FOXL2 directly interacts with the promoter of α-GSU, activating its expression (glycoprotein hormone α subunit which is common in Leutenizing hormone, Follicle stimulating hormone, and testicular hormone) [15].FOXL2 also directly interacts with steroidogenesis acute response (StAR) gene through its alanine-rich C-terminal region, to induce the inhibition of its transcriptional activity [16]. StAR protein enables the translocation of cholesterol from the cytoplasm to inside the mitochondria where pregnenolone is synthesized. Pregnenolone is the precursor for steroid hormones. Another function that FOXL2 plays in steroid hormone synthesis is that it is involved in the upregulation of CYP19, an aromatase involved in the conversion of androgens to estrogens in granulosa cells. CYP19 and FOXL2 have been observed existing together in goat ovaries [17]. Also, other factors involved in steroid biosynthesis like peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PPARGC1A) and NR5A2 are modulated by FOXL2.
Role in anti-oxidation
It is crucial to regulating the oxidative stress in the ovaries as ROS generation is high during ovulation [18-20]. PPARGC1A is not only involved in cholesterol metabolism, but it is also involved in the detoxification of reactive oxygen species (ROS). The expression of other components involved in the detoxification of ROS, namely immediate early response 3 (IER3) and Manganese superoxide dismutase (MnSD) are also upregulated by FOXL2 overexpression [21]. Therefore, FOXL2 also plays an important role in the detoxification of ROS. Also, upregulation of FOXL2 leads to cells pausing in the G1 phase where the damage induced to DNA by oxidative stress is repaired [18-20].
Role in Apoptosis
FOXL2 overexpression has been shown to induce the expression of BCL2-related protein A1 (BCL2A1) and tumor necrosis factor α – induced protein 3 (TNFAIP3) [21]. FOXL2 is also known to induce apoptosis using DEAD-box RNA Helicase DP103 as a coactivator [22, 23].
Role in Inflammation
It is involved in the regulation of PTGS2/COX2. COX2 is one of the two isoforms of cyclooxygenases which is involved in the synthesis of prostaglandins. COX2 also catalyzes the conversion of arachidonic acid to PGH2 which in turn is converted to other prostaglandins by synthases [24].
From the above-mentioned involvement of FOXL2 in different cellular pathways, it can be concluded that the fate of the cells in the ovary depends on the interaction of FOXL2 to the right set of proteins. As mentioned earlier, FOXL2 interacts with DP103 (also called DDX20 and Gemin-3), Steroidogenic factor 1 (SF1/NR5A1), estrogen receptors α and β (ESR1 and ESR2) and Smad3 [25, 26, 14, 27, 22, 23, 28-31].
When FOXL2 binds to SF1, it upregulates CYP19 aromatase expression and inhibits CYP17 expression which is mediated by unbound SF1. Therefore, FOXL2 plays a major role in balancing the levels of androgen (CYP17) and estrogen (CYP17). It has been observed that mutations in the SF1 lead to male-to-female sex reversal in humans [32] and FOXL2 missense mutation leads to female-to-male sex reversal in goats [33]. The interaction of FOXL2 with SF1 also upregulates Mc2r [34], which codes for an ACTH receptor required for steroidogenesis and adrenal gland development [1]. Upon interaction with ERα, it prevents the binding of ERα to PTGS2 promoter. Not much is known about the impact of the interaction of FOXL2 to ERα on another target of ERα [35].
FOXL2- A common observation in ovarian and non-ovarian type tumors
As discussed earlier, FOXL2 is generally found in the normal ovarian stroma. In addition, it is also found in ovarian neoplasms and non-ovarian neoplasms like pancreatic mucinous cystic neoplasms (PMCs), hepatobiliary cystadenomas (HBCs), and mixed epithelial and stromal tumor of the kidney (MEST). When the nucleus of cells of PMCs, HBCs and MEST was tested for FOXL2, 100% positivity was observed in PMCs and HBCs and 90% positivity was observed in MEST samples [36]. But this nuclear reactivity may not necessarily mean that there is a mutation in the FOXL2 gene which in turn has led to the tumor since FOXL2 was found in sex chord stromal tumors which lacked a mutation [37].
Sex hormone- cause for the neoplasm
Detailed analysis showed that these neoplasms are characterized by ovarian type stroma and they arise due to some history of hormonal therapy, obesity or heavy alcohol use. In the case of obesity, the insulin concentration is high which results in increased synthesis of androgen from ovaries and adrenals. The high levels of androgen precursors are converted to estrogen due to increased concentration of adipose tissue aromatases. Additionally, in obesity cases, there is an inhibition in the production of sex hormone binding globulin which makes the free estrogen levels to remain high [38].
In the case of alcohol abuse, the high intake of alcohol has been linked to increased levels of estrogen and androgen. Therefore, men who consume excessive alcohol develop gynecomastia, change in body hair patterns and testicular atrophy [39]. The last cause associated is hormone therapy where there is the administration of the exogenous hormone. This results in cancer of the breast, endometrial lining, and other estrogen-dependent cancers. Therefore, the common cause in all three cases is the increase in hormone levels.
One of the major observations regarding FOXL2 is that it serves as a good immune histochemical marker to identify ovarian type stroma in PMC, HBC, and MEST. Although the role it plays in these non-ovarian type tumors is yet to be established. [40].
FOXL2 in renal failure
In this paper, we will be concentrating on the effects on FOXL2 on renal failure and hence we will be speaking about the mixed epithelial and stromal tumor of the kidney (MEST). These tumors are rare and the case reports are isolated. They are called by the following alternate names leiomyomatous renal hamartomas, congenital mesoblastic nephroma in an adult, cystic hamartoma of the renal pelvis, solitary multilocular cysts of the kidney, multilocular renal cyst with Mullerian-like stroma, and adult metanephric stromal tumor [16]. It is more common among women and occurs in people of age ranging from 19-78. It is characterized by conspicuous abdominal or flank mass, flank pain and/or hematuria. The tumor is solid and cystic, tan to yellow in color with a clear shape from 2-24 cm in size [41]. It occurs in the renal hilum but does not infiltrate into the renal parenchyma. In rare cases, necrosis and calcification may be observed [42].
Cellular composition
The tumor is biphasic i,e., it has two types of cells – mesenchymal and epithelial. The mesenchymal component has spindle cells with varying degrees of differentiation, ranging from smooth muscle to fibroblastic to myofibroblastic cells interspersed with collagen. It resembles cystic nephroma or fibromatosis [29]. The epithelial component is found interspersed between the mesenchymal components. It varies from round and regular tubules to more complex tubule papillary structures with or without cystic dilatation. These are lined by cuboidal to the flattened epithelium (hobnail epithelium). Metastases are uncommon in cases of MEST. Until recently, malignant MESTs were not described. The malignancy can arise in either of the components. The malignant MESTs are characterized by increased cellularity, cytologic atypia, round to ovoid vacuolated nuclei with prominent nucleoli, and high mitotic rate. Rhabdoid, rhabdomyosarcomatous, and chondrosarcomatous components can also be observed sometimes [43, 44, 45].
Tumor markers
The epithelial components are positive for both low- and high- molecular- weight cytokeratin and Ulex europaeus [42, 46]. The mesenchymal components are positive for estrogen and progesterone receptors in most cases [47]. But the cells are negative for the receptor expression itself.
Common cause of MEST
An increase in estrogen and progesterone levels has been observed in MEST cases. Therefore, the increase in these steroid hormones is assumed to be the cause for MEST [48]. It has been already mentioned that FOXL2 gene plays an essential role in the production of estrogen and progesterone. A single case was identified which showed a translocation t(1;9) which is the cytogenetic cause of the MEST [49].
Differentiating MEST from other renal tumors
Unlike multicystic renal cell carcinoma, MEST lacks aggregates of clear cells [50]. Renal synovial sarcoma and MEST are almost similar except that MEST presents with subepithelial condensation of stroma (ovarian stroma) [51, 52, 53]. The characteristic ovarian stroma of MEST is not observed in rhabdoid tumor and the cystic and tubular hobnail epithelium is also not seen in the rhabdoid tumor [25]. Estrogen receptor positivity is a distinctive feature of MEST and it is used as a major immunohistochemical marker to differentiate from other tumors [50].
Material and methods
The FOXL2 protein sequence was downloaded from UniProt (Entry P58012) and subjected to BLAST to find if there were any homologs (Supplementary Table 1). Then, FOXL2 protein (accession number AAY21822.1) was modeled using PHYRE2 and I-TASSER. PHYRE2 predicts the structure using comparative modeling and I-TASSER does the structure prediction using ab initio modeling. Once the models were completed, they were subjected to further tests to check their quality. PROSESS and RAMPAGE were used to check the parameters of the predicted structure.
Results
BLAST results
According to the BLAST hit, almost all proteins are FOXL2 from various other organisms (Table 1). Only three of them are from other families of proteins namely human chorionic gonadotropin (hCG) and two unnamed proteins from Oncorhynchus mykiss.
Structure modeling
Since the crystal structure of the FOXL2 protein is not available, therefore, I tried to predict the model using in-silico tools and the structure was validated using PROSESS and RAMPAGE.described as follows.
I-Tasser model
I-TASSER predicted 5 different models and the best model was selected based on the one which had the highest C-Score. Figure 1 shows the selected model [40, 54, 34].
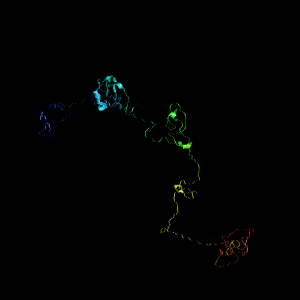 Fig. 1 Structure of FOXL2 predicted by I-Tasser.
Fig. 1 Structure of FOXL2 predicted by I-Tasser.
PROSESS result for the predicted model
When the I-TASSER model was subjected to PROSESS, the predicted model showed very poor quality. The non-covalent bond quality, torsion angle quality, and the covalent bond quality had scores of 2.5, 0.5 and 6.5 respectively. The non-covalent bond quality and torsion angle quality were the poorest features of among the three. This was because most of the parameters which contribute to these characteristics showed very high standard deviations. Even the flexibility of the predicted model was poor. As a result, the overall quality of the model itself was only 2.5 on a scale of 0 to 10.
RAMPAGE result for the predicted model
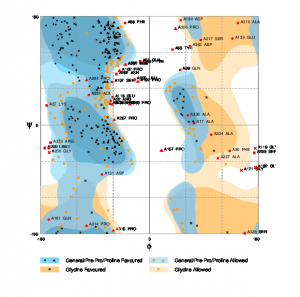
The Ramachandran plot was generated for the predicted model using RAMPAGE [55]. The plot is shown in Figure 2. According to the plot, the number of residues that should be in the favored region is only 58% of the total and the expected percentage is 98%. Similarly, the number of residues which should be present in the allowed region should be only 2% whereas this plot has 29.7% residues in the allowed region. The number of residues in the outlier region is 12.3%.
Fig. 2 RAMPAGE result for I-Tasser predicted model.
PHYRE2 model
The model predicted by PHYRE2 is shown in Figure 3 [56]. The structure was predicted using Interleukin Enhancer-Binding Factor 1. 25% of the sequence showed 100% confidence and the rest of the regions were deemed ‘disordered’.
Fig. 3 The structure of FOXL2 predicted by PHYRE2.
PROSESS result for the model
The model predicted by PHYRE2 was of moderate quality and not as poor as that of the I-TASSER model. The non-covalent bond quality, torsion angle quality, and covalent bond quality had scores of 5.5, 4.5 and 7.5 respectively. Similar to the I-TASSER model, non-covalent bond quality and torsion angle quality had the lowest scores among the three features. Only a few parameters of the two characteristics showed moderate standard deviations. The flexibility of the structure was also within the acceptable range. However, the overall quality score was still only 4.5 which is not very stable since the parameters were not up to satisfactory levels.
RAMPAGE result for the Model
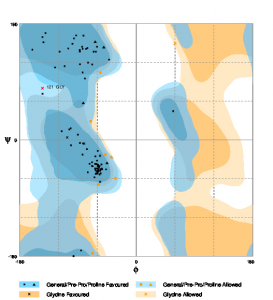
The Ramachandran Plot was generated using RAMPAGE [55]. Figure 4 shows the plot for the PHYRE2 model. The number of residues, in this case, is 90.6% and the number of residues in the allowed region is 8.3%. The number of residues in the outlier region is only 1%.
Fig. 4 RAMPAGE result for PHYRE2 predicted model
Conclusion
In conclusion, the structures predicted by both the software were of moderate to poor quality. Even though the structure predicted by PHYRE2 showed better characteristics and scores compared to I-TASSER, it is still not accurate enough. As a result, the model of FOXL2 has to be predicted using experimental techniques such as NMR or X-ray crystallography for a better understanding of the protein functioning.
Discussion
FOXL2 has a wide variety of biological applications but its role in kidney failure in not fully understood yet. The only common link between FOXL2 and MEST are the sex hormones estrogen and progesterone. This could probably mean that the gene for FOXL2 is somehow related to the increased concentration of these hormones which would lead to MEST and which in turn lead to renal failure. However, this has been the conclusion derived from reviewing the literature and detailed experimental studies are required to know the exact mechanism by which this is brought about.
Another obscure area is the structure of FOXL2 since not many attempts have been made to predict the structure of the protein. The in silico methods used in this study to predict the structure were not fruitful and resulted in unstable models. Therefore, experimental techniques are the solution to finding the accurate structure of FOXL2. Once the structure of the protein is known, one can get a better understanding of the protein which would consequently open a whole new perspective to the way it functions in so many different pathways.
References
- Cocquet et al. (2002) Evolution and expression of FOXL2 gene. J Med Genet, 39(12), 916–922, doi: 1136/jmg.39.12.916.
- Cocquet et al. (2003) Structure, evolution and expression of FOXL2 transcriptional unit. Cytogenet Genome Res, 101(3-4), 206–211, doi:10.1159/000074338.
- Baron et al. (2005) Foxl2 gene and the development of the ovary: a story about goat, mouse, fish and woman. Nutr. Dev., 45, 377–382, doi: 10.1051/rnd:2005028.
- Kaestner, K.H. et al. (2000) Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev., 14, 142–146, doi: 10.1101/gad.14.2.142.
- Treier, M. (1998) Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev., 12(11), 1691–1704, doi: 10.1101/gad.12.11.1691.
- Pannetier, M et al. (2003) Expression studies of the PIS-regulated genes suggest different mechanisms of sex determination within mammals. Cytogenet Genome Res, 101(3-4), 199–205, doi:10.1159/000074337.
- Zlotogora J. (1983) The blepharophimosis, ptosis, and epicanthus inversus syndrome: delineation of two types. Am J hum Genet, 35(5), 1020–1027.
- De Baere E et al. (2003) FOXL2 and BPES: mutational hotspots, phenotypic variability, and revision of the genotype-phenotype correlation. Am J hum Genet, 72(2), 478–487, doi: 10.1086/346118.
- Dollfus H et al. (2003) Sporadic and familial blepharophimosis- ptosis-epicanthus inversus syndrome: FOXL2 mutation screen and MRI study of the superior levator eyelid muscle. Clin Genet , 63(2), 117– 120.
- Barishak, Y. R. (1992) Embryology of the eye and its adnexae. Dev Ophthalmol, 24, 1–142 .
- Oley, C et al. (1988) Blepharophimosis, ptosis, epicanthus inversus syndrome (BPES syndrome). J med Genet, 25(1), 47–51 .
- Hersmus, R et al. (2008) FOXL2 and SOX9 as parameters of female and male gonadal differentiation in patients with various forms of disorders of sex development (DSD). J. Pathol., 215, 31–38, doi: 10.1002/path.2335.
- Loffler, K. A et al. (2003) Etiology of ovarian failure in blepharophimosis-ptosis-epicanthusinversus syndrome (BPES): FOXL2 is a conserved, early-acting gene in vertebrate ovarian development. Endocrinology, 144(7), 3237–3243, doi:10.1210/en.2002-0095.
- Ellsworth, B. S et al. (2003) The gonadotropin releasing hormone (GnRH) receptor activating sequence (GRAS) is a composite regulatory element that interacts with multiple classes of transcription factors including Smads, AP-1 and a forkhead DNA binding protein. Cell. Endocrinol., 206(1-2), 93–111.
- Ellsworth, B.S et al. (2006) FoxL2 in the Pituitary: Molecular, Genetic, and Developmental Analysis. Endocrinol., 20(11), 2796-2805, doi: 10.1210/me.2005-0303.
- Pisarska, M.D et al. (2004) Forkhead L2 is expressed in the ovary and represses the promoter activity of the Steroidogenic acute regulatory gene. Endocrinology, 145(7), 3424-3433, doi:10.1210/en.2003-1141.
- Pannetier, M et al. (2006) FoxL2 activates P450 aromatase gene transcription: towards a better characterization of the early steps of mammalian ovarian development. Mol. Endocrinol., 36(3), 399-413, doi:10.1677/jme.1.01947.
- Benayoun, B.A. (2009) Positive and negative feedback regulates the transcription factor FOXL2 in response to cell stress: evidence for a regulatory imbalance induced by disease-causing mutations. Mol. Genet., 18, 632–644, doi: 10.1093/hmg/ddn389.
- Benayoun, B.A et al. (2009) FOXL2: at the crossroads of female sex determination and ovarian function. Adv Exp Med Biol., 665, 207-26.
- Benayoun, B.A et al. (2009) The forkhead factor FOXL2: a novel tumor suppressor? Biophys. Acta, 1805, 1–5, doi: 10.1016/j.bbcan.2009.09.002.
- Batista, F et al. (2007) Potential targets of FOXL2, a transcription factor involved in craniofacial and follicular development, identified by transcriptomics. Nat. Acad. Sci. USA, 104, 3330-3335, doi: 10.1073/pnas.0611326104.
- Lee, K et al. (2005) Transcriptional factor FOXL2 interacts with DP103 and induces apoptosis. Biophys. Res. Commun., 336(3), 876–881, doi: 10.1016/j.bbrc.2005.08.184.
- Lee, M. B et al. (2005) The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Cell. Biol., 25(5), 1879–1890, doi:10.1128/MCB.25.5.1879-1890.2005.
- Smith, W.L., Dewitt, D.L. (1996) Prostaglandin endoperoxide H synthases-1 and -2. Adv. , 62, 167-215.
- Blount, A.L et al. (2001) FoxL2 and Smad3 coordinately regulate follistatin gene transcription. Biol. Chem., 284(12), 7631–7645, doi: 10.1074/jbc.M806676200.
- Corpuz, P.S et al. (2010) FoxL2 is required for activin induction of the mouse and human follicle-stimulating hormone {beta}-subunit Mol. Endocrinol., 24(5), 1037–1051, doi: 10.1210/me.2009-0425.
- Kim, S.Y et al. (2009) Foxl2, a forkhead transcription factor, modulates nonclassical activity of the estrogen receptor-alpha. Endocrinology, 150(11), 5085–5093, doi: 10.1210/en.2009-0313.
- Park, M et al. (2010) FOXL2 interacts with steroidogenic factor-1 (SF-1) and represses SF-1-induced CYP17 transcription in granulosa cells. Endocrinol., 24(5), 1024–1036, doi:10.1210/me.2009-0375.
- Turbiner, J et al. (2007) Cystic nephroma and mixed epithelial and stromal tumor of kidney: a detailed clinicopathologic analysis of 34 cases and proposal for renal epithelial and stromal tumor (REST) as a unifying term. Am J Surg Pathol., 31(4), 489–500, doi:10.1097/PAS.0b013e31802bdd56.
- Wang, D.S et al. (2009) Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Endocrinol., 21(3), 712–725, doi:10.1210/me.2006-0248.
- Yang, W.H et al. (2010) Synergistic activation of the Mc2r promoter by FOXL2 and NR5A1 in mice. Reprod., 83(5), 842–851, doi:10.1095/biolreprod.110.085621.
- Lin, L., et al. (2007) Heterozygous missense mutations in steroidogenic factor 1 (SF1/Ad4BP, NR5A1) are associated with 46, XY disorders of sex development with normal adrenal function. J.Clin. Endocrinol. Metab. 92(3), 991–999, doi: 1210/jc.2006-1672.
- Pailhoux, E et al. (2001) A 11.7-kb deletion triggers intersexuality and polledness in goats. Genet., 29(4), 453–458, doi:10.1038/ng769.
- Zhang, Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics, 9, 40, doi: 10.1186/1471-2105-9-40.
- Caburet, S., et al. (2011) The transcription factor FOXL2: At the crossroads of ovarian physiology and pathology. Molecular and Cellular Endocrinology, 356(1-2), 55-64, doi: 10.1016/j.mce.2011.06.019.
- Westerhoff et al. (2014) The expression of FOXL2 in pancreatic, hepatobiliary, and renal tumors with ovarian-type stroma. Human Pathology, 45(5), 1010–1014, doi: 10.1016/j.humpath.2013.12.015.
- Al-Agha, O. M et al. (2011) FOXL2 is a sensitive and specific marker for sex cord–stromal tumors of the Am J Surg Pathol, 35, 484-94, doi: 10.1097/PAS.0b013e31820a406c.
- Renehan, A. G et al. (2006) Obesity and cancer risk: the role of the insulin-IGF axis. Trends Endocrinol Metab, 17(8), 328-36, doi:10.1016/j.tem.2006.08.006.
- Sarkar, D. K et al. (2001) Role of estrogen in alcohol promotion of breast cancer and prolactinomas. Alcohol Clin Exp Res, 25(5), 230S-236S, doi: 10.1111/j.1530-0277.2001.tb02401.x.
- Yang, J et al. (2015) The I-TASSER Suite: Protein structure and function prediction. Nature Methods, 12(1), 7-8, doi: 10.1038/nmeth.3213.
- Pawade, J et al. (1993) Cystic hamartoma of the renal pelvis. Am J Surg Pathol., 17(11), 1169–1175
- Durham, J. R et al. (1993) Mesoblastic nephroma of adulthood: report of three cases. Am J Surg Pathol., 17(10), 1029– 1038.
- Jung, S. J et al. (2008) Mixed epithelial and stromal tumor of kidney with malignant transformation: report of two cases and review of literature. Hum , 39(3), 463–468, doi: 10.1016/j.humpath.2007.08.008.
- Nakachi, Y et al. (1997) Nucleotide compositional constraints on genomes generate alanine-, glycine-, and proline- rich structures in transcription factors. Mol Biol Evol, 14(10), 1042–1049.
- Svec, A et al. (2001) Malignant mixed epithelial and stromal tumor of the kidney. Virchows Arch., 439(5), 700–702.
- Truong, L. D et al. (1998) Adult mesoblastic nephroma: expansion of the morphologic spectrum and review of literature. Am J Surg Pathol., 22(7), 827–839
- Pierson, C. R et al. (2001) Mixed epithelial and stromal tumor of the kidney lacks the genetic alterations of cellular congenital mesoblastic Hum Pathol., 32(5), 513–520, doi:10.1053/hupa.2001.24323.
- Michal M, Syrucek M. (1998) Benign mixed epithelial and stromal tumor of the kidney. Pathol Res Pract., 194, 445–558.
- Comperat E et al. (2005) Benign mixed epithelial and stromal tumor of the kidney (MEST) with cytogenetic alteration. Pathol Res Pract., 200, 865–867, doi: 10.1016/j.prp.2004.05.004.
- Mohanty, S.K, Parwani, A.V. (2009) Mixed Epithelial and Stromal Tumors of the Kidney. Arch Pathol Lab Med, 133, 1483-1486.
- Argani, P et al. (1998) Detection of the SYT-SSX chimeric RNA of synovial sarcoma in paraffin embedded tissue and its application in problematic cases. Mod Pathol, 11, 65–71.
- Eble, J. N et al. (2004) Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. Lyons, France: IARC Press.
- Koyama, S et al. (2001) Primary synovial sarcoma of the kidney: report of a case confirmed by molecular detection of SYT-SSX2 fusion transcripts. Pathol Int., 51(5), 385–391, doi:10.1046/j.1440-1827.2001.01203.x.
- Yang J, Zhang Z (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Research, 43(W1), W174-W181, doi: 10.1093/nar/gkv342.
- Lovell, S.C et al. (2002) Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins: Structure, Function & Genetics., 50(3), 437-450, doi:10.1002/prot.10286.
- Kelley, L. A et al. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols, 10(6), 845-858, doi: 10.1038/nprot.2015.053.
- Biegel, J. A et al. (1996) Narrowing the critical region for a rhabdoid tumor locus in 22q11. Genes Chromosomes Cancer.,16(2), 94–105, doi: 10.1002/(SICI)1098-2264(199606)16:2<94::AID-GCC3>3.0.CO;2-Y.
- Chida, D et al. (2011) The role of glucocorticoids in pregnancy, parturition, lactation, and nurturing in melanocortin receptor 2-deficient mice. Endocrinology, 152(4), 1652–1660, doi: 10.1210/en.2010-0935.
- Cocquet J et al. (2003) Of compositional biases and polyalanine runs in man. Genetics, 165(3), 1613–1617.
- Nakagawa, T et al. (2004) Malignant mixed epithelial and stromal tumours of the kidney: a report of the first two cases with a fatal clinical outcome. , 44(3), 302–304, doi: 10.1111/j.1365-2559.2004.01782.x.
- Roy, A (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols, 5(4), 725-738, doi: 10.1038/nprot.2010.5.
- Uhlenhaut, N.H et al. (2009) Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell, 139(6), 1130–1142, doi: 10.1016/j.cell.2009.11.021.