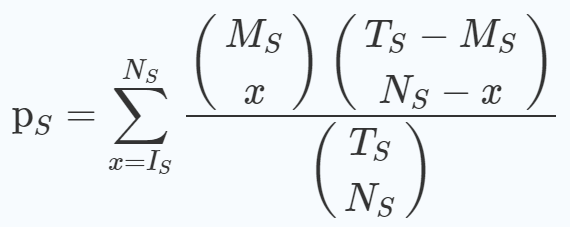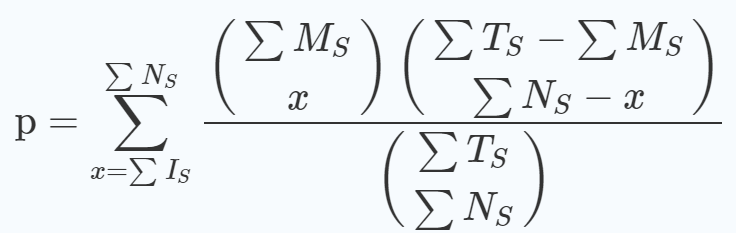small open reading frames (smORFs) that are less than 100 codons in length are considered significant. smORFs participate in a variety of biological processes such as muscle formation and contraction. Therefore, it is considered essential to identify the functions of these smORFs. For this purpose, a new tool called smORFunction has been developed [1].
smORFunction can predict functions of smORFs or microproteins in 265 models generated from 173 datasets. These datasets include tissues/cells, diseased, and normal. The webtool of smORFunction is available at https://www.cuilab.cn/smorfunction. Currently, it consists of 617,462 unique smORFs that are annotated by different tools SmPROT and sORFs.org. The data source can be downloaded from https://www.cuilab.cn/smorfunction/download.
How does smORFunction work?
It uses hypergeometric distributions for function prediction followed by the calculation of p-value for a set of functional genes using the following equation [1]:
Here, Ts =total background genes
Ms = genes present in a functional gene set
Is = screened genes to be correlated
A summarized p-value is calculated for all datasets using the following equation [1]:
smORFunction provides several options to search such as users can search using BLAST or exact mode or by coordinate in reference genome (GRCh37 or GRCh38). The performance and accuracy has been validated on 270 microproteins collected from literature and database. It can predict functions in at most 82 diseases (and normal), 48 tissues/cells. It also includes GO terms, KEGG pathways, and REACTOM pathways.
For further information, read here.
References
- Ji, X., Cui, C., & Cui, Q. (2020). smORFunction: a tool for predicting functions of small open reading frames and microproteins. BMC bioinformatics, 21(1), 1-13.