Pymol [1] is a molecular viewer that is mostly used for visualization purposes. Besides that, it can perform several other tasks such as mutagenesis. In this article, we are going to perform mutagenesis using Pymol [1].
Contents
Open Pymol by typing Pymol in a terminal or by double-clicking the shortcut or by launching the Pymol app. Now follow the steps explained below.
- Let’s open a protein structure first which we are going to mutate.
Go toFile --> Open --> Select an input PDB file. - Now go to
Wizard --> Mutagenesis. - Now pick a residue from the structure that you want to mutate.
- Look at the panel on the right. You will see ‘No Mutations’. Click on it and then select an amino acid to which you want to mutate.
- It will show you the structure of the amino acid to which you have mutated.
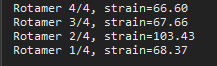
- It will generate a few rotamers that you can see in the lower right corner (Figure 1). Here is has generated 4 rotamers.
 Figure 1 Rotamers of the mutated residue.
Figure 1 Rotamers of the mutated residue. - Now cycle through the rotamers by using arrow keys. The red structures you see around the residue are the steric clashes. See the console as you move through the rotamers (Figure 2).
 Figure 2 Rotamers of the structure.
Figure 2 Rotamers of the structure. - After selecting a rotamer, click ‘Apply’ and then ‘Done’.
Now you can see the mutated residue in the structure and the sequence.
References
- The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.