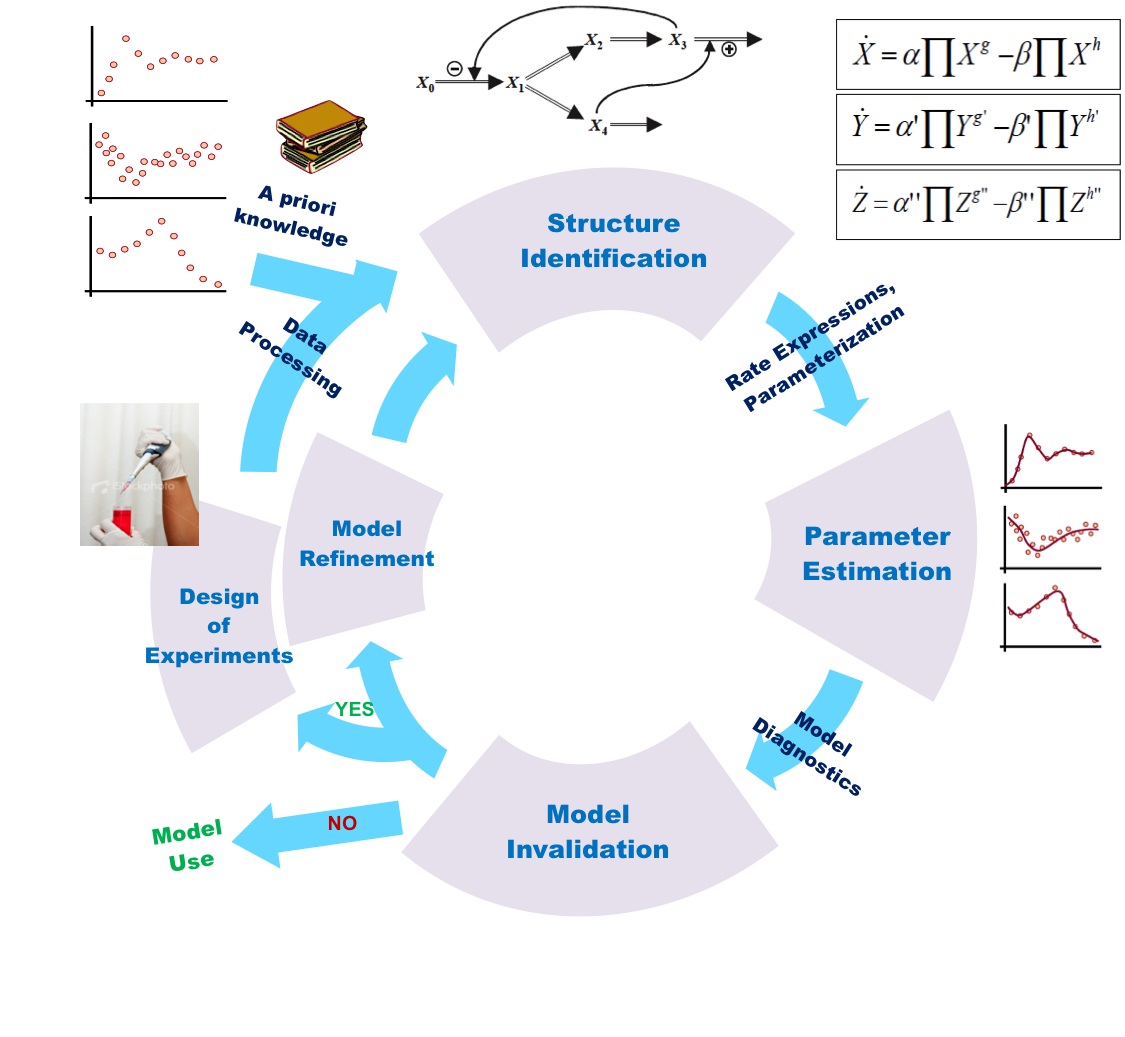
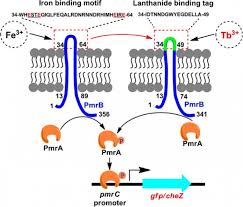
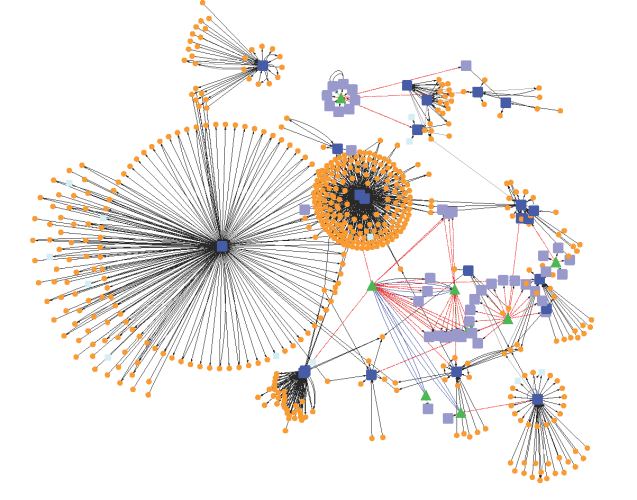
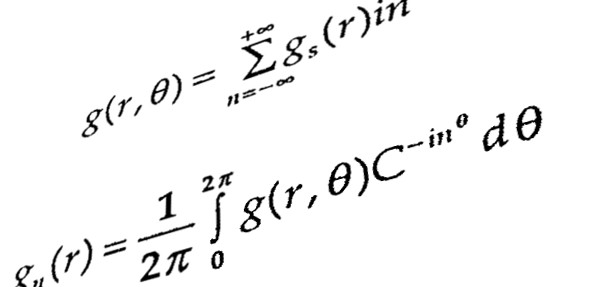In the previous section, mathematical modeling was exemplified by metabolic process and its biochemical regulation. It could also be done by signalling pathways and genetic regulatory process. At all cellular phase, one observe changing mode of a cell with effect from environmental factors. It is quite difficult to maintain cellular functions and reach to steady state. Thus, one needs to fix a range of parameters for all molecular reactions while going for mathematical modeling.
Identification of Model parameters:
Parameters for any equation in a model describe certain biochemical features of the components involved in reactions or pathways under study. For example, when modeling the dynamics of a metabolic network, the mathematical equations inferred from the processes must contain parameters that represent the kinetic features of the involved metabolic enzymes, as a number of reactions enzyme can perform within a given period of time (i.e., the rate constant). We must come across to these kinetic parameters prior to setting up well-defined systems of differential equations. Therefore, kinetic parameters for all the relevant reaction components can be experimentally determined. In practice, however, a number of kinetic parameters, even for otherwise well-investigated biological components (enzyme, proteins & hormones), still are not known, primarily because the reliable experimental data are lacking. It is very often that the kinetic parameters are measured but no experimental validation has been performed in wet lab (i.e. in vitro). In practice, enzymes behave similarly found in a cell. This creates a hurdle which is overcome by measuring the overall dynamics of the system being studied. Computational procedure have made them easier by providing appropriate estimation techniques to optimize parameter values by taking different multiple parameter set from the data set until they fit or get optimized for available experimental dataset. This method to is critically dependent on the quality of the dataset being validated, and therefore prediction made from such unreliable data will definitely lead to unreliable validated parameters and to a limited model of no use. In order to develop a simple network model for any biological process is awfully lagging behind mainly due to unavailability of high quality experimental data which is still a major focus in the field of systems biology.
It is important to mention that few biological process cannot be described using such simple models that are based on only concentration of molecules, ignoring the existence and importance of concerned components as the molecular movements adorns a significant impact on cellular mechanism. Due to the closely packing of the molecules in a cell, their thermal induction, and random movement from changing environmental conditions may cause the initiation of signal transmission that propagates across the cell and stops until reaches to its target. In order to account for such random effects, (i.e., stochastic) component must be incorporated into the equations of the model. For rare signaling molecules, of which lesser and fewer effect is observed, can be neglected. Whereas, molecules of which, rarest copies are existing in the cell must not be neglected and should be integrated into mathematical equations.
The next issue after optimization of parameter comes to be different time and distance scales of the components in integrated into a pathway. For example metabolism occurs in within seconds or minutes whereas genetic regulation takes longer (say it hours or even days) times to exert their effect or to express a particular gene induced by metabolic processes from a greater distance. It may be that signals (enzymes, protein, hormones) have to travel longer distance across the cell membrane via circulatory system of body fluids in between tissues. To overcome these different length and time scale, we can use multi-scale model to avoid complexity of the system.
Finally, it is important to assure that developed model is as good as assumption upon which it is based.