A network is a group of two more than two interacting components. The complex biological systems can be represented as computable networks that provide a unique way of analyzing the complex underlying mechanisms. For example, an ecosystem may be modeled as a network of interacting biotic and abiotic components and or a typical molecular mechanism can be modeled as a network of regulatory genes or proteins or even a network of protein can be constructed with different interacting amino acids. Further, a set of genes can be broken into a subset of small nucleic acids/genes interacting together to form a large network of regulating mechanism.
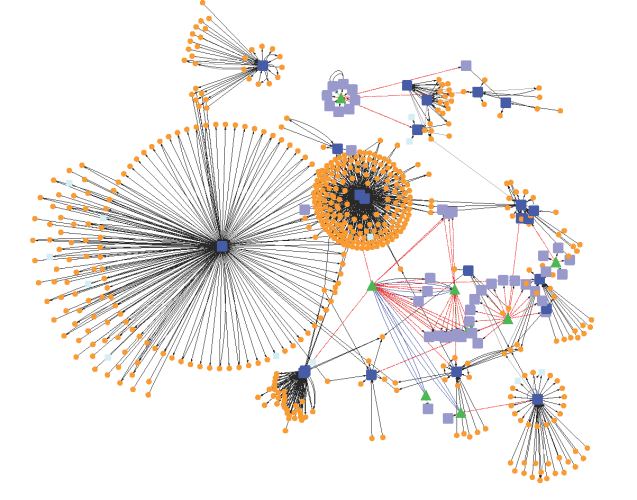
Even small components (atoms and or molecules) of a protein form an interacting network as carbon, oxygen, nitrogen, and sulfur. In every network, the connected components are considered as nodes and interactions between them are called as edges. In another language, a node represents a unit and an edge represents an interaction between the units. From a biological point of view, a node depicts a wide array of biological units. They may be anything from a tissue, cell, organ, cellular components, neurons, macromolecules to an individual atom. The most connected node i.e. node with the highest degree is called as Hub. Any Hub in a network is considered the most vital node as it has maximum links and becomes responsible for regulating other nodes. The overall functionality is affected if the hub gets disturbed.
Basically, a network is characterized by two of its properties viz. network degree and betweenness centrality. A degree of a node is the number of interactions/edges it shares with other nodes, while betweenness is a measure of centrality of a node that how central a node is, quantifying the number of times a node acts as a bridge along the shortest path between two other nodes in a network.
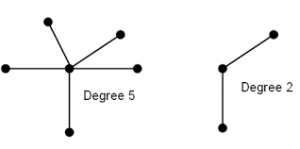
Fig: 1. The degree of nodes is represented in the graph as 5 of the central node while others have 1 as they are not linked with any node.
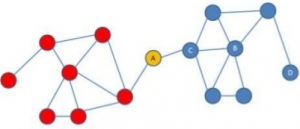
Fig: 2. The highly centered node is depicted in yellow color between two sub-sub graphs. Node A can exchange the information from both sides and can control the overall functionality of the network.
The highly centered node serves as a bridge among various portions of the network. An edge must pass through the node to reach other portion of the network. The importance of between centrality is varied for different networks like a social network, protein-protein interaction network in biology, gene-protein interaction and all regulatory networks. The centralized node is significant for passing the information from one sub-network to the other sub-network or from one portion to the other portion in a giant biological interaction network. Networks that we analyze are static whereas biological processes are greatly dynamic. There arrives a very special approach called mathematical modeling to deal with such a static graph to infer the useful information (refer to Introduction to Mathematical Modeling in the previous issue). Apart from, degree and betweenness centrality, other network properties include shortest path length, mean path length, diameter, density and average path length. They are referred to as topological properties of a network. Each one of them is significant in finding out the relationship between two nodes, flow/exchange of information, robustness, and functionality of a network, and can across to know about the overall function of biological systems i.e. robustness, redundancy, almost all biological networks hold a positive degree of the topological properties. Therefore, we can have a rough estimation of the function of a macromolecule (protein and or gene) in association with other connected molecules. The computational approach has made it easier to calculate the statics on topological properties. Soft-wares like Cytoscape and Gephi are widely used for visualizing and constructing biological networks.