Cancer
Tumor progression prediction by variability based expression signatures
Cancer has become a very common disease now a days, but the main reason of causing this is unknown up till now. Various reasons have been given and recent research says that improper sleeping patterns may also lead to cancer. Like cause of cancer is difficult to predict, similarly, its progression and prognosis is also very difficult. Despite of many advances in cancer treatment, early detection is still very difficult. While there have been many early cancer screening techniques, but are not realistic because of the lack of cost-effectiveness or requirement of invasive procedures. Genomic screening techniques are a promising approach in this area.
Gene expression signatures are commonly used to create cancer prognosis and diagnosis methods. Gene expression signature is a group of genes in a cell whose combined expression pattern is uniquely characteristic of a biological phenotype or a medical condition.
But only few of them were successfully able to utilize in clinics and many of them failed to perform. Since these signatures attempt to model the highly variable and unstable genomic behavior of cancer, they are unable to predict cancer. The degree of deviation in gene expression from the normal tissue, i.e., the hyper-variability across cancer types can be used as a measurement of risk of relapse or death. This gives rise to the concept of Gene expression anti-profiles. Anti- profiles are used to develop cancer genomic signatures that specifically takes advantage of gene expression heterogeneity. They explicitly model increased gene expression variability in cancer to define robust and reproducible gene expression signatures capable of accurately distinguish tumor samples from healthy controls.
Differentially variable genes= anti-profile genes
After employing many experiments regarding cancer anti-profiles, the results indicated that the anti-profile approach can be used as a more robust and stable indicator of tumor malignancy than traditional classification approaches.
The researchers’ hypothesis is that the degree of hyper-variability (w.r.t normal samples) is directly proportional to the tumor progression, i.e., degree of hyper- variability as measured with respect to the normal samples would increase with tumor progression. Corrada Bravo et al found out a way to derive a colon-cancer anti-profile for screening colon tumors by measuring deviation from normal colon samples. To create an anti-profile, they used a set of normal samples and tumor samples, probe- sets are then ranked by the quantity σj,tumor/ σ j,normal(where σj,tumor and σj,normal are the standard deviations among the tumor samples and normal samples, respectively, for probeset j) in descending order, and a certain number of probesets (typically 100) with the highest value are selected. Then they calculated the normal regions of each probe set and then the number of probe sets for which the expression lies outside the normal region was calculated to get an anti-profile score of the sample.
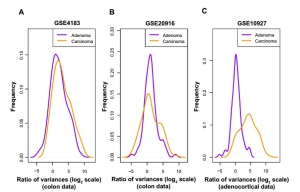
To test their hypothesis, they obtained two publicly available microarray datasets with normal, adenoma, and cancer colon samples. By studying these datasets, they plotted the distribution of variance of cancer/adenoma samples to variance of normal samples ratio (in log2 scale) for these probe sets on the other dataset (Fig. 1A and B).
Fig.1 Among probes that exhibit higher variability among cancers than among normals, the degree of hypervariability observed is related to the level of progression. (a) Distribution of variance ratio statistic {log2 σ σ2tumor ÷ σ2normal} for colon dataset (Gyorffy et al; GSE4183) from anti-profile computed using another colon dataset (Skrzypczak et al; GSE20916). (B) Distribution of variance ratio statistic for Skrzypczak et al colon dataset from anti-profile computed using Gyorffy et al colon dataset. (C) Distribution of variance ratio statistic for adrenocortical data (Giordano et al; GSE10927) for universal anti-profile probe sets.
Both adenoma and cancer samples show higher variability than normals (region to the right of x = 0), while cancer samples show higher hypervariability than adenomas. This suggests that hypervariability is a stable marker between experimental datasets and that specific selection of hypervariable genes across cancer types and the anti-profile method can be extended to model tumor progression. These studies showed that Gene expression anti-profiles capture tumor progression.
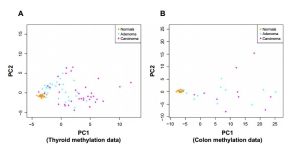
DNA methylation is one of the primary epigenetic mechanisms for gene regulation, and is believed to play a particularly important role in cancer. High levels of methylation in promoter regions are usually associated with low transcription. Cancer has loss of sharply methylation levels which is associated with increased hypervariability in gene expression across multiple tumor types. They applied the anti-profile scoring method to DNA methylation data from thyroid and colon samples, where for each tissue type, normal, adenoma and cancer samples were available. Figure 2 shows the distribution of adenoma and carcinoma samples against normal samples on a principal component plot, showing the presence of the hypervariability pattern in methylation data: the normal samples cluster tightly, while the adenomas show some dispersion and the carcinomas show even greater dispersion. Since these behaviors are present for both colon and thyroid data, it again reinforces their notion that the anti-profile approach has wide application for classification in cancer.
figure 2. Anti-profiles applied to methylation data: first two principal components of (A) thyroid methylation data and (B) colon methylation data.
Conclusion:
The anti-profile approach is more suitable for cancer prognosis. It can robustly predict the tumor progression and prognosis based on the variability in the gene expressions. The results presented above also confirms that gene expression signatures based on hyper-variability can be highly valuable.
References:
Wikum Dinalankara and Héctor Corrada Bravo Center for Bioinformatics and Computational Biology, Department of Computer Science and UMIACS, University of Maryland, College Park, MD, USA

Cancer
Cancer: From the Eyes of Mathematical/Systems Biology
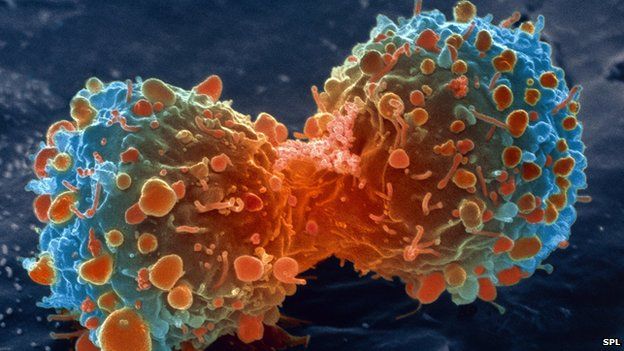
The month of November has just arrived with its generic glimpse of winter. We welcome this month with an evergreen and hot topic of cancer research. This time we intend to introduce you to an old research topic with a new vision…..
Cancer being an ailment with no remedy of full confidence has been pursued as a career by a lot of researchers. A cell biologist says it is an uncontrolled proliferation (increase in number by division and growth) of cells, molecular biologists call it a mutant variety of some biomolecules forcing a cell to commit such an uncontrolled cell division cycle. But, how does a Systems Biologist see such kind of a problem? Let us try to pursue it in a different way.
Proteins if are not assigned some name based on their function or structure, scientists mark them according to their molecular weight, e.g. p53, p200, p19 etc. Scientists have proven an abnormally high expression of p53 protein in Cancerous cells/tissues. p53 protein is actually the reason behind those other proteins which regulate the cell cycle and makes it to divide in to two as a normal scenario, p53 also helps in the manufacture of its inhibitor named Mdm2 protein. In any case of mutation in p53, that leads the failure of abnormality recognition by p53, doesn’t lead to increase in p53 and consequently Mdm2, p21 and other p53 regulated proteins. And thus, the division of abnormal cells continues indefinitely and causes Cancer.
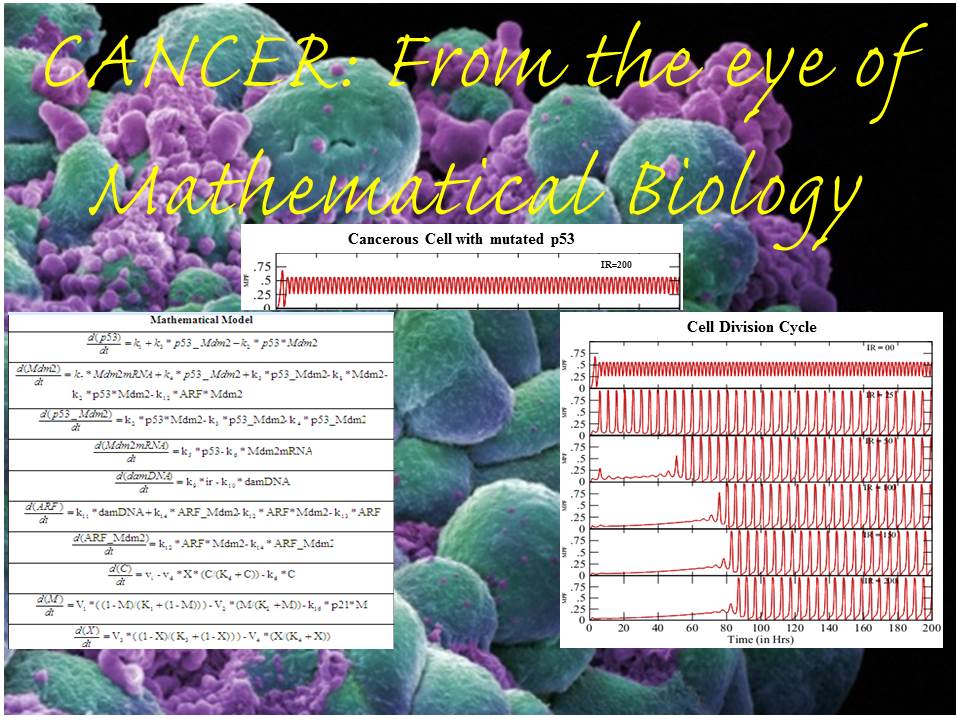
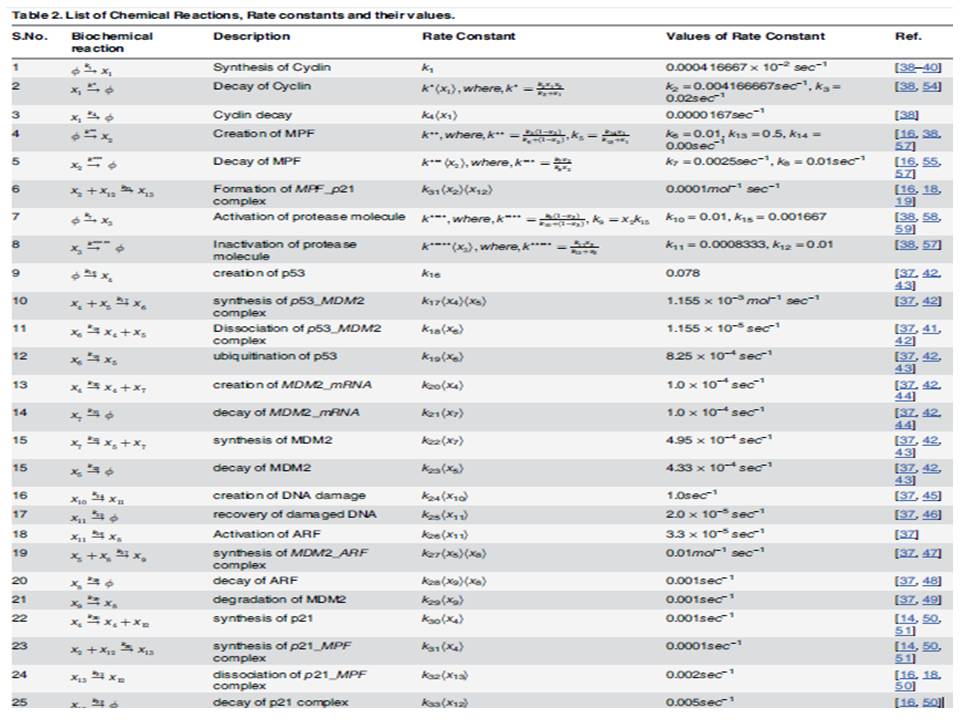
From a Mathematical Biology perspective, systems biologists form some ordinary differential equations that look like a mathematical formula. These mathematical formulae are actually nothing else than the representative of chemical reactions and their combinations occurring inside a cell. As in our previous blogs (by Fozail Ahmad), we have mentioned about how to combine the chemical reactions in a shape of Ordinary Differential Equations (ODEs) and about how we follow Zero-Order chemical kinetics (reaction rate doesn’t depend on any participating chemical), First-Order chemical kinetics (reaction rate depends on only one participating chemical) and Second-Order chemical kinetics (reaction rate depends on two or more participating chemicals) to form the equations. In addition to that, I would like to mention that there are some reactions which occur with the help of some biomolecular machineries. These machines (enzymes) just help the reactions to occur, but do not take part in it themselves and thus affect the reaction in a different form of kinetics as described by the combined work of German Scientist of Biochemistry Leonor Michaelis and Canadian Scientist of Physics Maud Menten in 1913.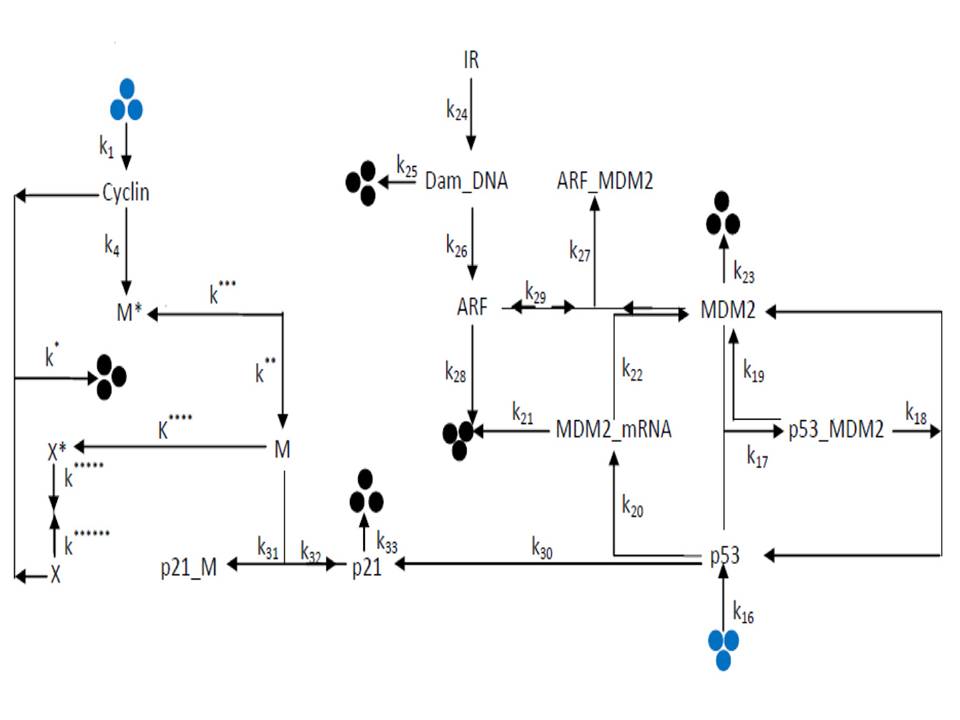
So, in a normal cell, when p53 senses the danger and signals the Cell by increasing p21 to combine with PCNA (Proliferating Cell Nuclear Antigen – An enzyme that helps in cell division) it stops the cell division. This type of cell cycle division has been shown in one of the diagrams mentioned below, while for the mutated case of p53 where it can not sense the cellular damage and thus divides normally is also shown in one of the images above.
We have also mentioned a combined picture, which shows a referral of how different stages of Mathematical Biology looks like. These figures are in special contrast to Cancer cells and normal cells.
Reference: Alam MJ, Kumar S, Singh V, Singh RKB (2015) Bifurcation in Cell Cycle Dynamics Regulated by p53. PLoS ONE 10(6): e0129620. doi:10.1371/journal.pone.0129620
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0129620



You must be logged in to post a comment Login