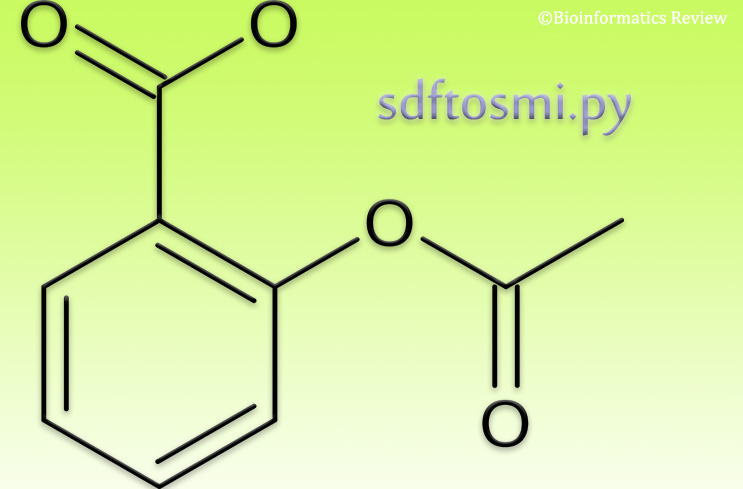
You can obtain SMILES of multiple compounds or ligands in an SDF file in one go. Here, we provide a simple Python script to do that.
Let’s say, we have a file in SDF format consisting of molecular structures called ‘ligands.sdf’. In order to perform some other operations, you can convert SDF to SMILES. It will not only make it easier to process but also reduce the size of the file, especially when you are dealing with millions of compounds.
Availability
The script is available on GitHub under the package ‘cheminformatics‘.
Usage
The Python script named sdftosmi.py will take the input (SDF file) filename as an argument and will provide output in the ‘smiles.txt‘ file. This file will contain SMILE corresponding to each compound in the SDF file. Therefore, you may have several duplicates because a SMILE can represent different molecules.
Run the script as shown below:
$ python3 sdftosmi.py ligands.sdf