Gathering of Dynamic/Kinetic information
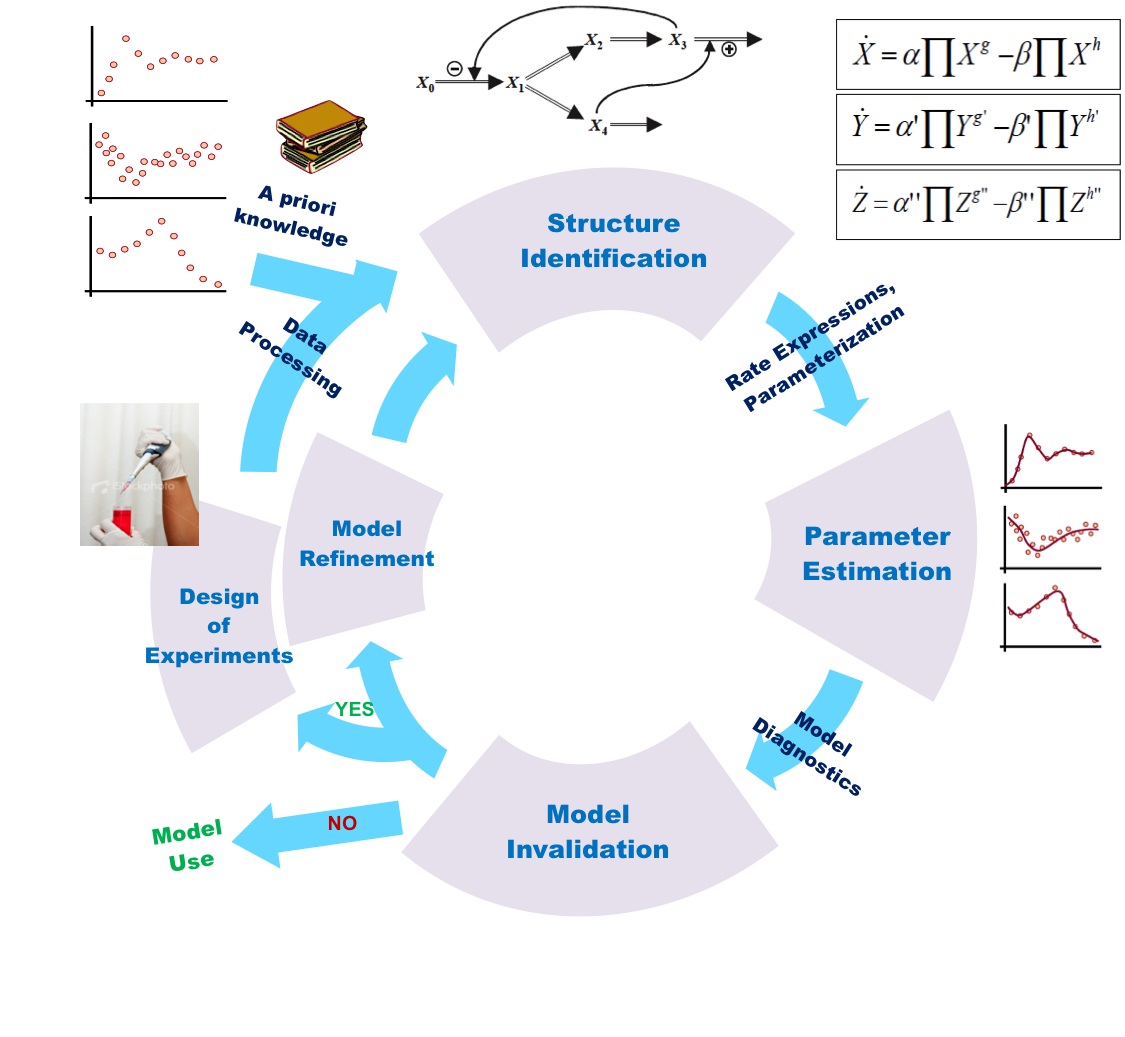
In the previous section you might have noticed that modelling biochemical process requires calibrated set of fine parameters which fit into and across the set of chemical/reactant species (gene/protein/molecule) involved in the process.
Question arises, where do we collect data from? And what are the standard criteria for determining parameters? Basically, for a researcher, it is necessary to know the source of the data first and then how to manipulate to get relevant information for modelling. Source of the data can be chosen depending upon the requirement of experimental design. For modelling, data can be taken in the form of gene-gene interaction, Gene expression (micro-array) and gene-protein interaction. Basically, interaction and expression do not simply reveal the dynamic/kinetic values of the system and therefore need to be manipulated for further implication.
In order to obtain the parameter values for analysing the kinetic behaviour of biochemical processes, we simply investigate into expression data (gene/protein) at different transcriptional and translational level that enable us to frame-out the comprehensive structural biochemical pathway. This can be done by accounting the following methods:
- Genome (complete set of genes) analysis at transcription level through DNA sequencing & genotyping
- Transcriptome (all mRNA) analysis at translation level using microarrays
- Proteome (entire protein) analysis at cellular level (reaction between proteins and other molecule in cell) using Mass spectroscopy and 2D gel electrophoresis
- Metabolome (total metabolites and their intermediate) analysis at cell level, interaction metabolites and regulator using 13C labelling and NMR techniques
- Interactome (all interacting molecule) analysis by yeast-2 hybrid screen & TAP techniques
(Ligand: TAP- Tandem affinity purification, NMR- Nuclear magnetic resonance)
Above mentioned techniques are collectively referred to as Omics. They provide us structural and dynamic data that is used to generate mathematical formula representing observable reactions, followed by development of mathematical model and comprehensive pathway of biological system. These tentative models allow us (as mentioned in part-1) to observe the effect of a stimulus on specific signalling pathway, perturbation in cellular activities and gene expression level etc.
Omics are characterized by a number of features. First, they allow researcher for analyses on different molecular levels such as gene, protein and metabolites level. These different molecular levels sometimes show asynchronous behaviour—that is, although some metabolite such as glucose is higher in a cell and corresponding enzymes are lower than sugar to catalyse the reaction or vice versa. Asynchronous behaviour is an indication of complex regulatory mechanism. Therefore, it is crucially important to evaluate the degree of synchronization of all cellular level. Second, Omics are highly parallelized. This means all genes/mRNA (read-outs in sample) can be studied simultaneously rather than having to perform separate experiments focusing on individual genes. This parallelization also allows researcher to compare the degree of expression results for the same gene and to have an interaction between resultant proteins. Third, they are very standardized and therefore needs very high automation computing, providing scientist with a large number of samples at time. In the process, after collecting huge data, most relevant information is picked up and then processed further for final analyses.
Entireties of techniques in an Omics are very important in the sense that they generate numerical data based upon which we are able to develop structural pathway for mimicking the real picture of biological system and then to represent in the form of mathematical model.
( →Continue to part 3 )