Molecular dynamics (MD) simulation has become an important methodology in research covering systems consisting of millions of atoms. We have provided several articles on MD simulation including the GROMACS [1] installation and performing MD. In this article, we will learn about “QwikMD” a plugin in NAMD [3], and VMD [4].
QwikMD allows users to set up the MD simulation of macromolecules in a few minutes. The steps are explained as follows.
- Open terminal (Ctrl+Alt+T) and type
$ VMD. (If you are using Windows, then open a command prompt by typing ‘cmd’ in the search box and then type>VMD). - Go to
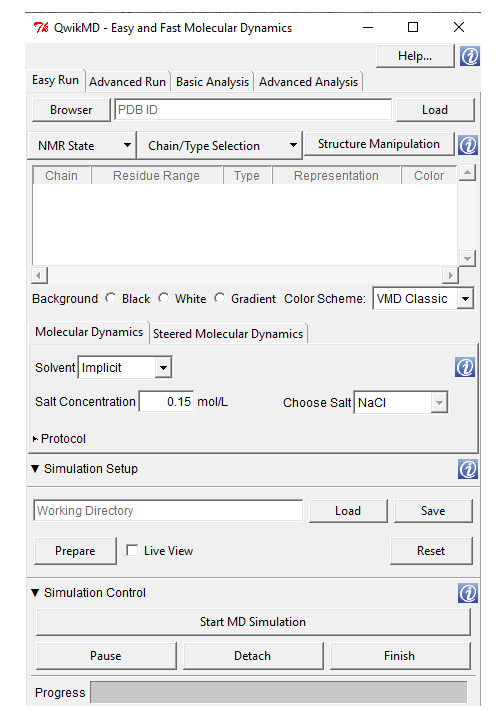
Extensions --> Simulate --> QwikMD. A window will appear as shown in Fig. 1.
Fig. 1 A screenshot of QwikMD window launched from VMD
3. Now, load your protein structure either by typing or by browsing, then Click 'Load' .
It allows users to either simulate in an explicit solvent by simulating all atoms of the solvent or in an implicit solvent by adding a dielectric constant to the electrostatic calculation. For more information regarding this, click here. We are performing a simple MD and we will be using implicit solvent method.
4. Remove oxygen atoms from water molecules.
Click on 'Chain/Type Selection' and deselect the water molecules 'A and water'.
After that, you can see the oxygen atoms (red) in the VMD display will disappear.
5. Select the ‘Solvent’ and ‘Salt concentration’. Here, it is ‘implicit’ and default salt concentration (0.15 mol/L).
6. Now, move to the section ‘Simulation Set up’.
Check the 'Live View' checkbox for Live simulation and click 'Prepare'.
During this step, autopsf function is called to generate files required for MD simulation including NAMD configuration files. You will have to select a folder to save the input file named ‘.qwikmd‘.
After that, you will notice two folders will be created in the working directory: setup and run. All files created during the set up will be stored in the setup folder and those files that will be required for running MD simulation will be stored in run folder.
7. Move to another option “Protocol”.
Check the 'Equilibration' and 'MD' checkboxes and enter particular 'Temperature' and 'Simulation time'.
Here, the temperature was set to 27 C (300 K) and the simulation time was set to 1ns.
8. Run the simulation
Click 'Start MD Simulation'--> Select the number of CPU cores to be used to perform the simulation.
It might show an error (” Error connecting to localhost on port 3000“) until a connection is established between NAMD and VMD. The live simulation will be continued to run in the background until you abort it.
That’s it. Wait for the simulation to finish.
References
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1, 19-25.
- Ribeiro, J. V., Bernardi, R. C., Rudack, T., Schulten, K., & Tajkhorshid, E. (2018). QwikMD-Gateway for Easy Simulation with VMD and NAMD. Biophysical Journal, 114(3), 673a-674a.
- Phillips, J. C., Zheng, G., Kumar, S., & Kalé, L. V. (2002, November). NAMD: Biomolecular simulation on thousands of processors. In SC’02: Proceedings of the 2002 ACM/IEEE conference on Supercomputing (pp. 36-36). IEEE.
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: visual molecular dynamics. Journal of molecular graphics, 14(1), 33-38.