Covalent Docking
How to perform covalent docking using AutodockFR (ADFR)?
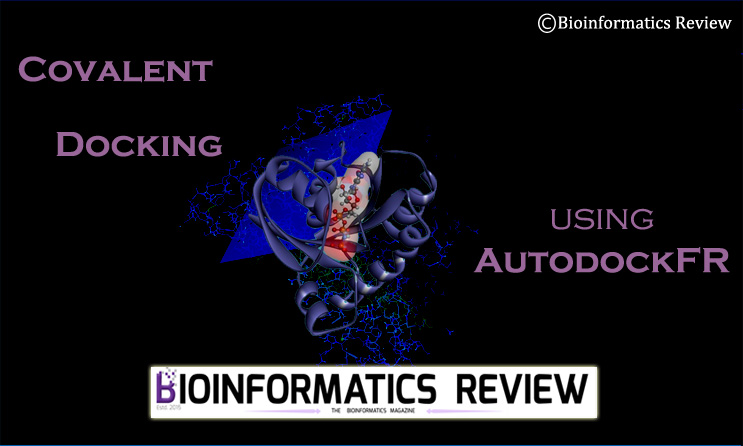
Docking is performed in different ways depending upon the requirements. In some cases, it is required to dock a covalent ligand with the receptor. In this article, we will perform docking of a covalently bound ligand with a receptor using AGFR [1] and ADFR [2]. You can install these tools from the MGLTools2 package. For the installation guide, click here.
In a covalently bound ligand, there are three atoms shared between the receptor and the ligand. It includes a common anchor atom in the receptor that connects with the ligand’s atoms. You can see both connected atoms in the PDBQT files of both the receptor and the ligand as well. In this tutorial, we are docking 4-Chlorobenzoyl-CoA Ligase/Synthetase (PDB ID: 3CW9) bound to 4-chlorophenacyl-CoA.
Prepare PDBQT files
Prepare the PDBQT file of the receptor and the ligand using Autodock tools.
Preparation of PDBQT file of ligand
You can randomly optimize the ligand’s conformation as well as its positions and orientation in the crystal structure. Then save as the PDBQT file as shown below.
- Open
AutoDock Tools --> Ligand --> Input--> Open
It will ask you to select your ligand, we will go to the folder where we have saved our ligand’s .pdb file and click “ligand.pdb”.
- Click
Ligand --> Torsion Tree --> Detect Root.
It will show the torsion angle on the ligand from where it can be rotated.
- Click
Ligand --> Output --> Save as PDBQT
We can rename the ligand, but we will use the same name as before and will name it as “ligand.pdbqt” and save it in the same folder.
Preparation of PDBQT file of receptor
Let’s save the receptor file as “protein.pdb“. Now, prepare the PDBQT file of receptor protein using the same procedure as used for simple site-specific docking except for a few steps.
Let’s prepare a simple PDBQT file.
- Open AutoDock Tools, click
File --> Read Molecule --> select protein.pdb. - We will delete water molecules from the protein as they can make unnecessary bonds with the ligand. Click
Edit --> Delete water. - We will add polar hydrogens in order to avoid any empty group/ atom left in the protein. Click
Edit --> Add Hydrogens --> Polar only - We will save this file as .pdbqt, click
Edit Grid --> Macromolecule --> Choose --> select the “protein.pdb” --> OK. It will ask for a folder to save, then save it as “protein.pdbqt“, in the same folder where the pdbqt file of the ligand was saved. - Locate residues involved in making covalent bond. For details on how to locate residues, read this article.
- Remember, in covalent docking, there is no need for coordinates to define a grid box. The ligand is placed in the grid box using the covalent bond.
Prepare target file
Note down the PDB serial number of the atoms forming a covalent bond present in the PDBQT file of the receptor. In this case, 1593 and 1596.
Generate the target file
We will use AGFR to calculate the affinity maps. It helps to specify the grid box position for docking. You can either use the command-line or GUI for that. Here, we will be using the command line.
$ agfr -r protein.pdbqt -b user 28.565 6.329 6.985 25 25 25 -c 1593 1596 -t 1591 -x A:CYS164 -o protein_affmaps
Here, -r represents the receptor PDBQT file.
-b defines the box manually.
-c defines the serial numbers of atoms forming a covalent bond in the receptor.
-t represents the anchor atom defining the covalent attachment and is used to calculate the torsion angle of the covalent bond.
-x argument limits the parsing of receptor to covalent residues only as in some cases, covalent residues can create bonds with the receptor other than the covalent attachment.
-o defines the output filename.
It will output the mentioned file with .trg extension.
Analyze the generated target file
You can see all information in the target file using the following command:
$ about protein_affmaps.trg
It will display all necessary information including maps, pocketMode, box, and receptor.
Perform covalent docking
Now, you can dock the ligand with the receptor using the following command:
$ adfr -l ligand.pdbqt -t protein_affmaps.trg --jobName covalentdocking -C 1 2 3 --nbRuns 8 --maxEvals 100000 -O --seed 1
Here, -l defines the ligand.
-t defines the generated target file containing affinity maps.
-maxEvals define the number of evaluations of the scoring function. You can modify these values according to your requirements.
The output files are named according to the job name specified in the above command (--jobName covalentdocking). After the processing finishes, it will generate three output files:
- ligand_covalentdocking_summary.dlg
- ligand_covalentdocking_out.pdbqt
- ligand_covalentdocking.dro
The output analysis will be explained in the upcoming tutorial.
References
- Zhao, Y., Stoffler, D., & Sanner, M. (2006). Hierarchical and multi-resolution representation of protein flexibility. Bioinformatics, 22(22), 2768-2774.
- Ravindranath, P. A., Forli, S., Goodsell, D. S., Olson, A. J., & Sanner, M. F. (2015). AutoDockFR: advances in protein-ligand docking with explicitly specified binding site flexibility. PLoS computational biology, 11(12), e1004586.







[email protected]
January 24, 2022 at 7:46 pm
I followed your procedure its quite similar with (https://ccsb.scripps.edu/adfr/tutorial-covalent/) software tutorial. The issue is if I use prepared tutorial input material for the exersise, it give -6.3kcal/mol binding energy (descent exersise). They use the ligand with ash-propyl chain (why i dont know, not even mentioned in any tutorial).
If I prepare the ligand and protein as per given process, it depict positive binding energy and do not dock.
For your kind consideration i am elaborating the process.
1. I downloaded the protein (3c9w) from protein data-bank
2. Imported it into AutoDock tools, removed water and chain B
3. Extracted the ligand and prepared with considering torsion then save it in pdbqt format.
3. Now a) i used apo protein.That is removed co-crystalline covalently bound ligand and satisfy valency b) protein with covalently bound ligand (used in original tutorial) and generate trj file.
4. during .trj file generation i carefully inspect atom numbers. Tey are identical
5. After docking i end up with positive energy with different docking pose.
Kindly help me to figure it out.