Evolution
Tutorial-I: Functional Divergence Analysis using DIVERGE 3.0 software
DIVERGE is a tool to detect functional divergence between member genes of a protein family based on (site-specific) shifted evolutionary rates after gene duplication or speciation [1]. Since this tool contains several steps and offers a lot to discover, therefore, we are going to split its tutorial into parts. In this article, we will perform some initial steps in this software and will explain their significance.
You can easily download and install the latest version of DIVERGE 3.0. It works on Windows. To start the software, double click on the application in the Diverge directory.
1. Input
You need an alignment file in .aln or .fasta format as an input file.
- Click on the ‘Load Alignment‘ button and open your input file. It will display complete alignment on the DIVERGE window.
- You can also input a reference protein structure of any of the proteins you have in your alignment. For that, click on the ‘Load Structure‘ button and open the PDB file. It will display the structure in a new window.
2. Clustering
After inputting the alignment and the structure, you need a tree for clustering. You have two options, either you can input your phylogenetic tree of the aligned sequences or you can make one.
- Click on the ‘Load Tree‘ button and open a tree file. The extension of the tree file should be .tree to be recognized by the DIVERGE software.
- Or make a new tree by clicking on ‘NJ-Tree Making’. You can make the tree using three methods: p-Distance, Poisson, and Kimura.
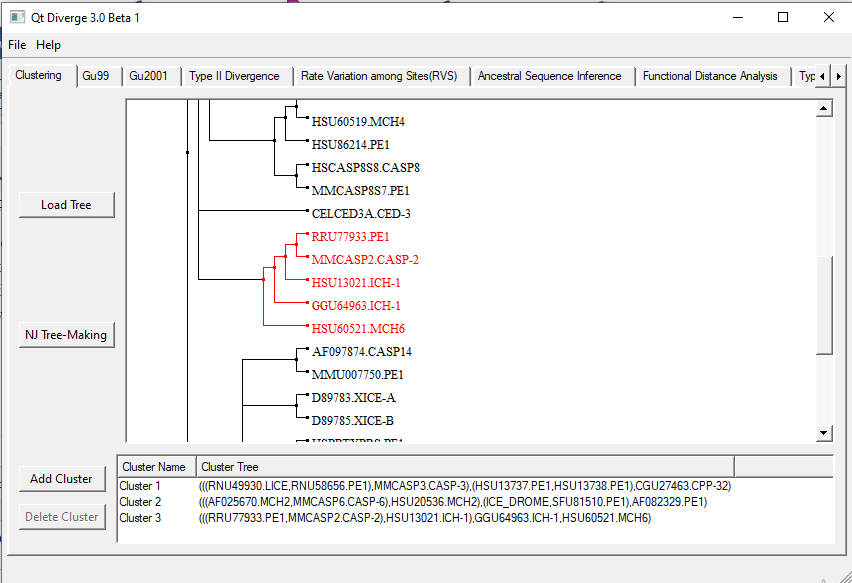
A tree will appear on the screen in the same Clustering tab. Now, cluster the sequences.
2.1. Clustering sequences
Remember each cluster must have at least three sequences.
- Look at the tree and select clusters by clicking on the branches or nodes. The selected branches/node will appear red.
- After selecting a cluster, go down to the bottom panel. There you will see another button namely, ‘Add Cluster’.
- Click on ‘Add Cluster’ and name it as you wish.
- Similarly, add more clusters. It will look like this (Figure 1):
Figure 1. Sequence clustering in DIVERGE 3.0.
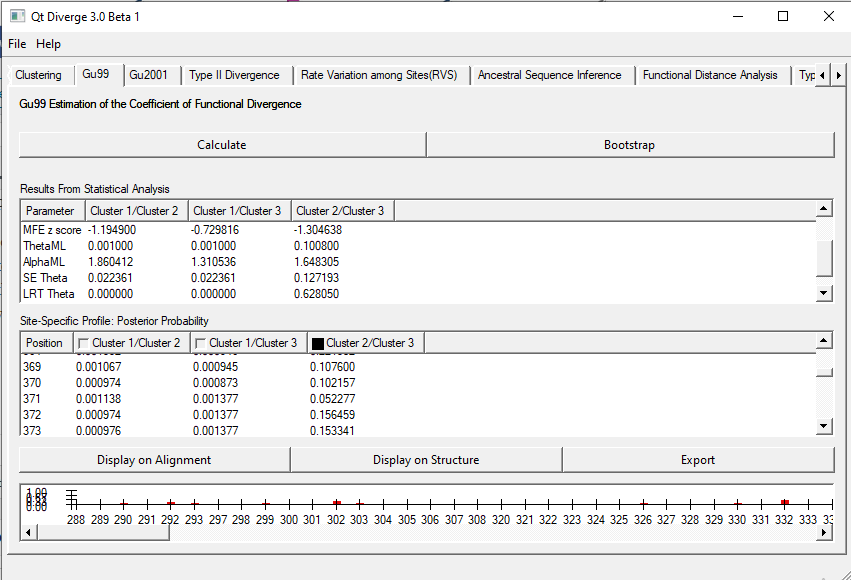
3. Gu99 test
This test estimates the coefficient of functional divergence. To calculate this, click ‘Calculate’. You can also set the bootstrapping value. by default, it is 100.
It will show the sites that have undergone functional divergence. The site-specific values would be given for a set of two clusters. For example, Cluster1/Cluster2, Cluster1/Cluster3, and Cluster2/Cluster3. The results will look like as shown in Figure 2.
Figure 2. Gu99 test on DIVERGE 3.0
You can also visualize these sites on the alignment as well as on the loaded structure. If there is no significant posterior probability, then it won’t show anything on the plot visible in the bottom panel. Additionally, you can export the values in a text document.
4. Gu2001 test
It is the new version of the previous test. You can use either of them. Follow the steps mentioned in the Gu99 test. Remember this test will work only if you have loaded the tree with branch lengths.
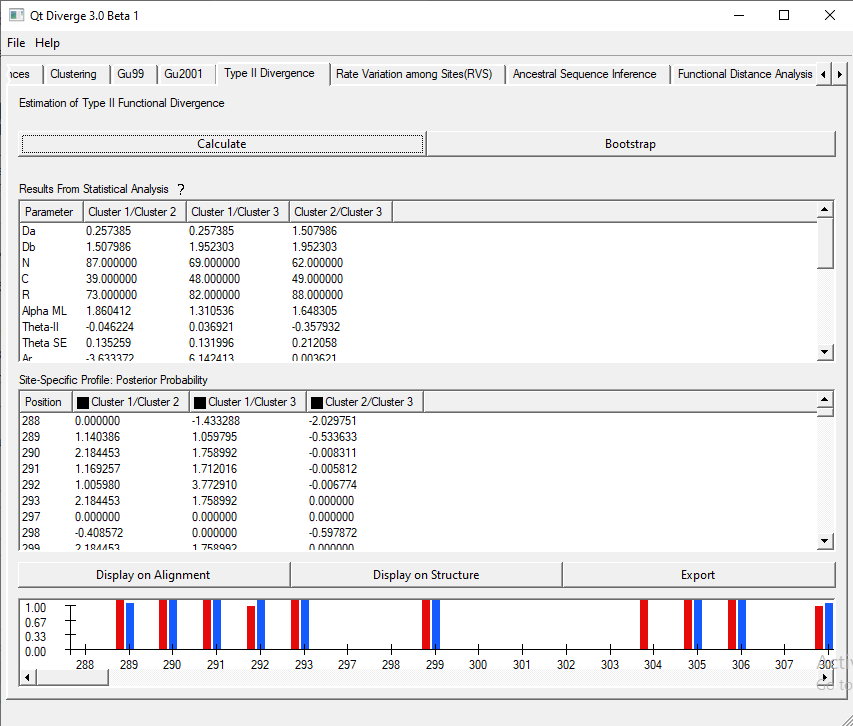
5. Type-II Divergence
This test calculates the Type-II functional divergence coefficient (ӨII). ӨII identifies the radical amino acid changes at some sites caused by the rapid evolution [2].
- To perform this test, click ‘Calculate’ and wait for the results. The results are shown in Figure 3.
Figure 3. Type-II Functional Divergence analysis using DIVERGE 3.0.
- You can see the sites in single or all cluster combinations. These sites are also visible on the alignment and the structure.
Further analysis will be explained in the upcoming article.
References
- Gu, X., Zou, Y., Su, Z., Huang, W., Zhou, Z., Arendsee, Z., & Zeng, Y. (2013). An update of DIVERGE software for functional divergence analysis of protein family. Molecular biology and evolution, 30(7), 1713-1719.
- Gu, X. (2006). A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Molecular biology and evolution, 23(10), 1937-1945.
Evolution
Tutorial-II: Functional Divergence Analysis using DIVERGE 3.0 software

This is a continuation article of our previous post: Tutorial-I: Functional Divergence Analysis using DIVERGE 3.0 software. In our last tutorial, we explained Type-II functional divergence. In this article, we will explain the rest of the evolutionary analysis using DIVERGE 3.0 software [1]. (more…)
Evolution
Update: Selection analysis on spike glycoproteins of SARS-CoV
In our last article, we mentioned the selection pressure analysis of SARS-CoV-2 spike glycoproteins. Now, we have analyzed spike glycoprotein sequences of SARS-CoV. No single purifying selection site was found in SARS-CoV spike glycoproteins as was revealed from our last analysis of SARS-CoV-2 spike glycoproteins. (more…)
Evolution
Update: Selection analysis on spike glycoprotein of SARS-CoV-2
We have recently conducted a selection pressure analysis on SARS-CoV-2 spike glycoprotein sequences. The analysis revealed not even a single site showing evidence of purifying selection but episodic diversifying selection on three sites. Out of these three sites, two sites are potentially relevant (Fig. 3). (more…)



[email protected]
December 4, 2021 at 9:19 am
Hello,
I am trying to use this software, but I don’t know how to get the file with the .tree suffix. The tree file I currently have has a suffix of .newick.
Dr. Muniba Faiza
December 5, 2021 at 4:56 pm
Hi,
Basically, .tree is the Newick format. You can simply edit the extension to the .tree file.
[email protected]
December 6, 2021 at 7:10 am
Yes,I have edit that.But for some reason the program still reported bug.I am checking my ducuments.
If I make tree by program,the program will run.But the NJ-tree made by program are different from my already-made-tree.So maybe there are some wrong with my tree file.