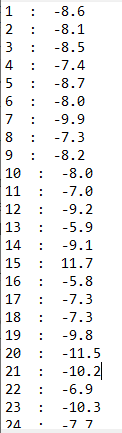In one of our previous posts, we provided a Python script for the virtual screening analysis of Autodock Vina. This script analyzes all log files obtained from the docking of multiple ligands to a receptor and provides the binding affinities for top poses from each file. In this article, we are publishing a new Python script for the virtual screening analysis of SMINA [1].
SMINA is a fork of Autodock Vina that improves scoring and minimization. It provides a single log file as output which contains all the binding affinities of the docked ligands. However, it does include the name or identity of the docked ligands. Therefore, it is difficult to identify which binding affinity belongs to which ligand. For this purpose, an SDF file as output does the trick. You can use sdsorter to fetch the binding affinities including the name or identity of the associated ligands. Here is a tutorial to install sdsorter on Ubuntu.
SMINA log file can be useful in conditions where you need only binding affinities of the ligands regardless of their identities. One such example is the AUC-ROC analysis, calculation enrichment factor (EF) values, and so on.
sminalog_analysis.py script fetches all binding affinities of the top poses of each ligand giving a serial number to each one of them from the SMINA output log file.
Usage
Download the file and save it in the directory where you have kept all log files. If you save it elsewhere, don’t forget to provide the full path to this file. It will run on Linux as well as on Windows.
$ python3 sminalog_analysis.py
After running the above command, you will see the directory path where you have kept the file along with its name. A few seconds later, it will display “Done! The result is provided in the output.txt file”. A new file will be created namely, ‘output.txt’, in the same directory. This file will contain the final results.
Example:
Say, you have docked 3000 ligands from a database and want to get the binding affinities of the top poses of each of these ligands. Then run the above command and wait for the output file. The output will look like this:
Availability
The script is freely available to download from the Bitbucket account of Bioinformatics Review.
NOTE:
- This script screens for the log files containing the word ‘log’ in their filenames, e.g., “log_docking.txt” or “log_3YPI.txt”, or “2yp1_log_docking.txt” etc.
How to cite this script:
Faiza M., Abdullah T. (2021). sminalog_analysis.py – A Python script to fetch top binding affinities from SMINA log file. 8(6):page 16-18. The article is available at https://bioinformaticsreview.com/20210611/sminalog_analysis-py-a-new-python-script-to-fetch-top-binding-affinities-from-smina-log-file
For any queries, contact me at muniba@bioinformaticsreview.com.
References
- Koes, D. R., Baumgartner, M. P., & Camacho, C. J. (2013). Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. Journal of chemical information and modeling, 53(8), 1893-1904.
Further Reading
vs_Analysis.py: A Python Script to Analyze Virtual Screening Results of Autodock Vina
Virtual Screening Methodology for Structure-based Drug Designing