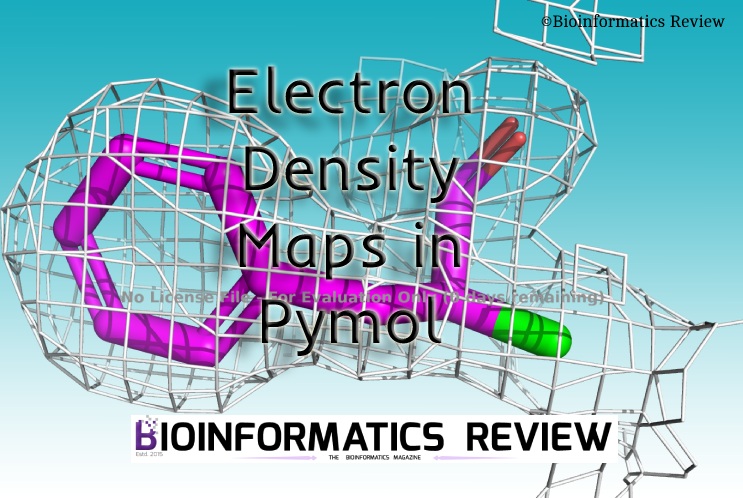
Electron density maps are available for most of the protein structures in PDB. Therefore, in this article, we are using PDB to generate electron density maps in Pymol.
Electron Density Map of whole protein
Follow these steps to generate an electron density map of the whole protein structure:
- Open Pymol. We will use the command line.
- Set the view. This is optional, if you don’t want to set the view then skip to the next step.
> set orthoscopic
> set line_width, 5 - Download the structure and electron density data from PDB.
> fetch 1db1, async=0
> fetch 1db1, type=2fofc, async=0 - Now, create the map at 2.5-angstrom carve and map level of 1.06.
> isomesh 1db1_map,1db1_2fofc, 1.06, poly, carve=2.5
You can adjust the map level. But if you increase this number then it will become shorter on display. - Select a residue to focus on.
> orient i. 279
It will zoom in on the selected residue. - You can set the colors as well.
> color magenta, e. C
> color yellow, e. N
Electron Density Map of selected residue
Follow these steps to generate an electron density map of selected residue:
- Open Pymol. We will use the command line.
- Set the view. This is optional, if you don’t want to set the view then skip to the next step.
> set orthoscopic
> set line_width, 5 - Download the structure and electron density data from PDB.
> fetch 1db1, async=0
> fetch 1db1, type=2fofc, async=0 - Select a residue and extract it.
For example, select residue 279 –> right side panel –> ‘sele‘ –> ‘A‘ –> ‘extract object‘.
Rename the object if you want. Here, we are using the default name (obj01). - Hide the rest of the protein. This step is optional, if you wish to display the rest of the protein then skip to the next step.
Go to –> Right side panel –> ‘1db1‘ –> ‘H‘ –> ‘hide everything’. Or just simply click on the name of the structure (1db1). - Now, create the map at 2.5-angstrom carve.
> isomesh mesh_ligand_carved, 1db1_2fofc, selection=(obj01), carve=2.5 - Select a residue to focus on.
> orient i. 279
It will zoom in on the selected residue. - You can set the colors as well.
> color magenta, e. C
> color yellow, e. N
References
- The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.