The Microbial Pan Genome is the union of genes shared by genomes of interest. This term was first used by Medini in 2005.
Since then, microbial genome data has been enormously increased, so to study processes such as selection and evolution, the construction of pan genome of species is required. But construction of pan genome from the real data available is very difficult and would not be accurate due to fragmented assemblies, poor annotation and also the contamination,i.e., microbial organisms can rapidly acquire genes from other organisms. Therefore, Andrew J. Page et al have developed
a new method to generate the pan genome of a set of related prokaryotic isolates and named the tool as ‘Roary’. It deals with thousands of isolates in a feasible time.
How Roary Works?
One annotated assembly per sample is input in the Roary from which coding regions are extracted and converted in to protein sequences, and all the partial sequences are removed and pre clustered using CD-HIT (a fast program for clustering and comparing). This produces a reduced set of protein sequences.These reduced sequences are compared all-against-all with the help of BLASTP with a user defined percentage sequence identity (default 95%). Now, by using conserved neighborhood genes, homologous groups are split in to true orthologs. Finally, a graph is constructed showing the relationships of the clusters based on the order of occurrence in the input sequences.
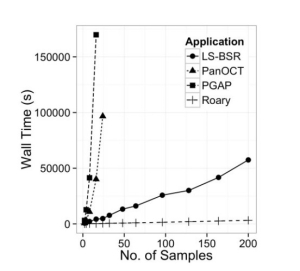
Fig.1 Effect of dataset size on the wall time of multiple applications.
That’s how the orthologous genes of prokaryotes can be easily identified and the microbial evolution can be well studied. It is done on a large scale covering a large data set to analyse the pan genomes of prokaryotes. Other tools have also been made earlier than Roary for the same purpose,namely, PanOCT and PGAP, but Roary is more fast, heuristic and most feasible tool among them.
Note:
An exhaustive list of references for this article is available with the author and is available on personal request, for more details write to muniba@bioinformaticsreview.com.