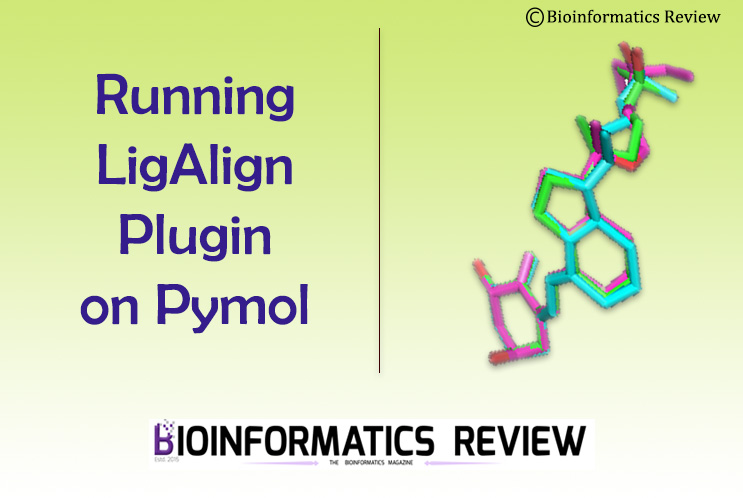
Running a plugin on an old version of Pymol [1] can give you multiple errors that are not easy to troubleshoot. For example, LigAlign plugin [2] runs on an old version of Pymol. Previously, we explained how to install LigAlign on Pymol. In this article, we will run the LigAlign command on Pymol.
After a successful installation of the LigAlign plugin, you will see “LigAlign v1.00 loaded” on the Pymol console. But when you try to run this plugin, it still gives you errors including,
“ExecutiveLoad-Error: Unable to open file ‘2bxa'” OR
“NameError: global name ‘CmdException’ is not defined”
Therefore, you will have to perform some additional steps to make it run. They are explained below.
Preparing input
Let’s take three PDB structures as input. You can use any complex structure. We are using 2BXA, 2BXB, and 2BXC.
- Download these structures in any format, PDB or CIF. We are using the CIF format here.
- Copy these downloaded structures in the same directory where the ligand_alignment.py script is located. This script is present inside the ligalign directory. If you haven’t installed ligalign yet, then visit this link.
Running LigAlign
- Open Pymol. Change to the directory where the ligand_alignment.py script is located. For example,
PYMOL> cd /home/user/Downloads/ligalign
It will display the path on the console. - Now open those structures. Go to File –> Open –> Select the structures.
- Run the following command in the Pymol console.
PYMOL> ligalign 2bxa.cif, 2bxb.cif, 2bxc.cif - You will see aligned ligands in the display and RMS values in the console.
If this doesn’t work then follow the steps shown below:
- Open Pymol. Change to the directory where the ligand_alignment.py script is located. For example,
PYMOL> cd /home/user/Downloads/ligalign
It will display the path on the console. - Now fetch the structures.
PYMOL> fetch 2bxa
PYMOL> fetch 2bxb
PYMOL> fetch 2bxc - Run the following command in the Pymol console.
PYMOL> ligalign 2bxa.pdb, 2bxb.pdb, 2bxc.pdb - You will see aligned ligands in the display and RMS values in the console.
References
- Heifets, A., & Lilien, R. H. (2010). LigAlign: flexible ligand-based active site alignment and analysis. Journal of Molecular Graphics and Modelling, 29(1), 93-101.
- The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.