Data Mining
PcircRNA_finder: Tool to predict circular RNA in plants

The non-coding circular RNAs (circRNA) play important role in controlling cellular processes. circRNAs are covalently bonded continuous closed loops which originate from the exonic region, known as exonic circRNA, but they can also arise from the intronic and the intergenic regions. CircRNAs can serve as a miRNA sponge [1,2] and are capable of enhancing the transcription of their host genes [3,4]. A new rRNA-depleted high-throughput RNA-Seq technology has revolutionized the discovery of circRNA in most of the species such as the mouse, human, rice and Arabidopsis [5,6].
There are several circRNA prediction tools available such as find_circ and CIRCexplorer, which were primarily developed using the databases of animals or humans [2,7,8-10]. Since these prediction methods are used to predict cricRNA for animal genomes and there is a large difference between the genome of animals and plants, therefore, these method can not be used to predict the cricRNA of plants, otherwise, the results would not be accurate [11,12]. Li et al.,(2016), has developed a software known as “PcircRNA_finder” to predict the circRNA in plants genome [12].
Workflow of PcircRNA_finder:
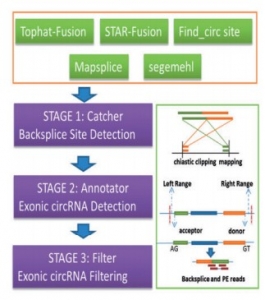
Fig.1 Workflow of pcircRNA_finder [12]
This software is basically designed to predict the exonic circRNA consisting of three modules (Fig.1):
- Catcher
It maps the paired-end reads and collect all backsplice sites on the basis of presently available fusion detection methods such as STAR-Fusion [13], Tophat-Fusion [14], find_circ [2], Mapsplice [15] and segemehl [16]. This gives backsplice sites and false positive sites which are filtered by the other module.
- Annotator
It annotates the candidate exonic backsplice sites on the basis of the available gene annotation [12]. It has been proved by recent studies that the backsplicing sites of circRNAs are flexible and their alternative splicing of circRNAs is prevalent [9,17]. Li et al., (2016) allowed the 5-bp flanking as many of the alternative splice sites occurred near the canonical splicing sites [12].
- Filter
This module functions for the quality control of the predicted circRNAs and requires at least one of the two kinds of splicing signals [18,19] –
a) a U2 based spliceosome (usually with a consensus sequence of GT-AG and GC-AG) and
b) a U12-based minor spliceosome (usually with a consensus sequence of AT-AC)
This module works as follows :
i) Creates a pseudoRef file which holds all the flanking sequences of chiastic backsplice sites.
ii) Maps raw reads to it.
iii) Confirm the backsplice sites.
PcircRNA_finder has been found to be more accurate after testing with a benchmark dataset by Li et al., (2016) [12].
For further reading, click here.
REFERENCES:
- Hansen,T.B. et al. (2013) Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384–388
- Memczak,S. et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495, 333–338.
- Li,Z. et al. (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol., 22, 256–264.
- Zhang,Y. et al. (2013) Circular intronic long noncoding RNAs. Mol. Cell, 51, 792–806.
- Lu,T. et al. (2015) Transcriptome-wide investigation of circular RNAs in rice. RNA, 21, 2076–2087.
- Ye,C.Y. et al. (2015) Widespread noncoding circular RNAs in plants. New Phytol., 208, 88–95.
- Pan,X. and Xiong,K. (2015) PredcircRNA: computational classification of circular RNA from other long non-coding RNA using hybrid features. Mol. Biosyst., 11, 2219–2226
- Salzman,J. et al. (2013) Cell-type specific features of circular RNA expression. PLoS Genet., 9, e1003777
- Szabo,L. et al. (2015) Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol., 16, 126.,
- Zhang,X.O. et al. (2014) Complementary sequence-mediated exon circularization. Cell, 159, 134–147
- Ye,C.Y. et al. (2015) Widespread noncoding circular RNAs in plants. New Phytol., 208, 88–95
- Li Chen , Yongyi Yu , Xinchen Zhang , Chen Liu , Chuyu Ye and Longjiang Fan. PcircRNA_finder: a software for circRNA prediction in plants. Bioinformatics, 2016, 1–2 doi: 10.1093/bioinformatics/btw496
- Dobin,A. et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21.
- Kim,D. and Salzberg,S.L. (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol., 12, R72
- Wang,K. et al. (2010) MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res., 38, e178.
- Hoffmann,S. et al. (2014) A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol., 15, R34.
- Starke,S. et al. (2015) Exon circularization requires canonical splice signals. Cell Rep., 10, 103–111
- Reddy,A.S. et al. (2013) Complexity of the alternative splicing landscape in plants. Plant Cell, 25, 3657–3683
- Staiger,D. and Brown,J.W. (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell, 25, 3640–3656
How to cite this article:
Faiza, M., (2016) miRNA targets and their functions. Bioinformatics Review, 2(9):page 9-13. The article is available at http://bioinformaticsreview.com/20160910/pcircrna_finder-tool-to-predict-circular-rna-in-plants/
Data Mining
Bioinformatics data mining: an introduction

Bioinformaticians handle a large amount of data: in TBs if not in gigs thus it becomes important not only to store such massive data but also making sense out of them. In this article, I will talk about what is data mining and how bioinformaticians can benefit from it.
What is data mining?
Data Mining is the process of discovering a new data/pattern/information/understandable models from ha uge amount of data that already exists. It is sometimes also referred to as “Knowledge Discovery in Databases” (KDD). (more…)



You must be logged in to post a comment Login