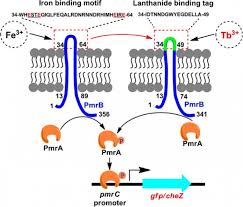
The genomic complexity and unknown functions of proteins/genes in Mycobacterium tuberculosis (Mt) has triggered an in-depth study of the entire genome to explore factors responsible for influencing Mt’s behaviour at molecular level. To set the stage of infection, to establish itself in the host’s defending environment, to cause the pathogenicity by overcoming the immune system and to escape out from any assailable host attack, this TB causing pathogen has developed a well-embodied system known as two-component system (TCS) that constitutes two proteins, universally designated as sensor protein and response regulator protein.
The basic function of these proteins is to sense environmental signals and respond accordingly. After interaction with suitable stimulating ligands, sensor protein, histidine kinase binds and hydrolyzes ATP, catalysing the auto-phosphorylation of a conserved Histidine residue and producing a high energy phosphoryl group. The phosphate is then transferred to the associated receiver protein known as response regulator at conserved Aspartic acid residue generating a high-energy acyl phosphate. Once phosphotransfer reaction has taken place, the response regulator gets activated, allowing it to carry out its specific function. In most of the cases, activated sensor kinase modulates the transcription of DNA at a specific binding site located in target genome at promoter region. The total effect is change in global gene expression that aids pathogen to respond in the initial signal sensed by histidine kinase. There are eleven such TCS in the pathogen. The primary task of such system is to control the expression of specific genes at specific time in response to the environmental conditions hence contributing to the growth of pathogen inside host. Since each of the TCS is obligated with distinct function, they are involved in orchestrating most of the gene regulatory processes. Out of eleven, only eight TCS have been studied comprehensively letting others to be scavenged by further genomic analysis of Mt.
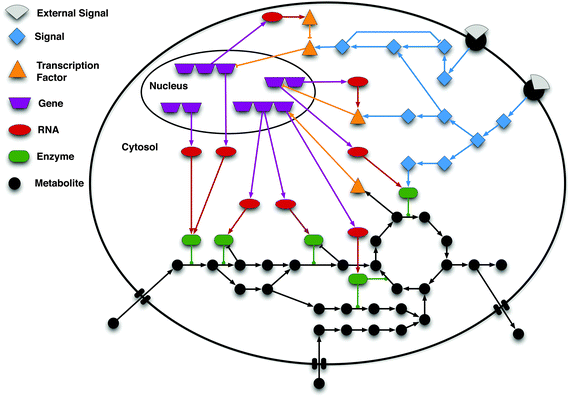
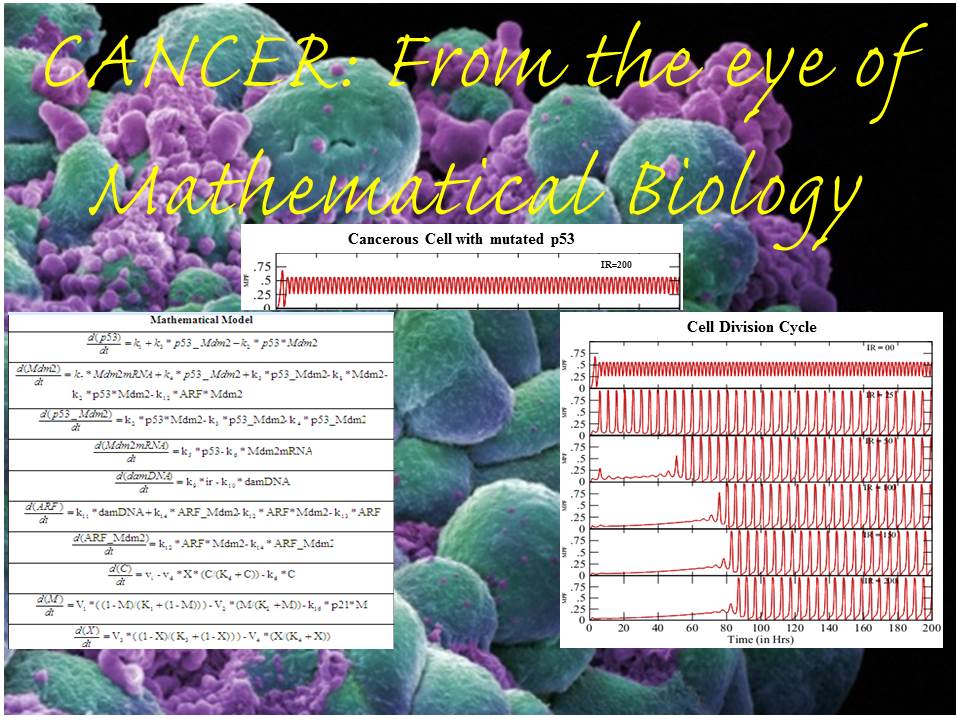
Interdisciplinary relevance : The systematic understanding of biological phenomena and demonstration of such microscopic processes have been subjected to a number of sophisticated experimental procedure in order to develop the deterministic or stochastic approaches that are skilled in unfolding real molecular system. Biological modeling and simulation are among those of biochemical annotating methodologies using wet lab data and understanding the scenario of real biological mechanism. Systems biology opens a new area to analyse the raw data generated through wet lab experimentations by various modes of characterization and evaluation by mathematical modeling, simulations & network analyses as the sole implications into any biological issue. Two-component systems for their critical contributions in bacterial pathogenicity have provided us with new concepts for comprehending molecular mechanism which are yet to be explored. Limitations have been raised for it’s behaviour and activation so far as the exact regulatory mechanism is concerned. Application of mathematical model and simulation over the regulatory behaviour would testify the real global association of TCS with entire genomic expression showing how this pathogen becomes so potentially virulent? Another important question that can be raised is at what level of gene activation the pathogenicity is rampant making host unimmunized? The scavenging initiative of all two-component systems would bring the molecular biology, chemistry, mathematics and network biology together to unfold the gene regulatory scenario of Mycobacterium tuberculosis in an exclusive manner.







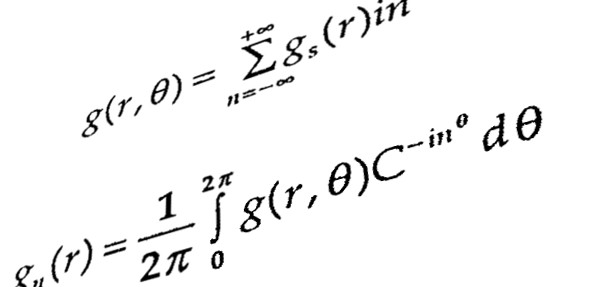
Thank You Fozail for the very informative and interesting article
your welcome Sir