Databases
McPAS-TCR: A database of TCR sequences associated with pathology and antigens
T-cells, also known as T-lymphocytes, play important role in the cell-mediated immune system. T-cells consist of a T-cell receptor (TCR) at their surface which recognizes the foreign and self- origin antigens. These TCRs composed of two chains, namely, TCRα and TCRβ chains which are produced by a random process of DNA rearrangement [1,2]. These TCR repertoires are highly diverse ensuring an effective immune system which cannot be even captured in a blood sample [3].
Recent research has reported the mapping of TCR repertoires using high-throughput sequencing [4-6] and most recently a manually curated database for TCR sequences is developed which are associated with pathology and the antigens [7].
Tickotsky et al., [7] have tried to link the TCR with its respective antigen to which it binds and recongnizes along with the pathological condition it causes such as cancer, infections, and other diseases in humans and mice.
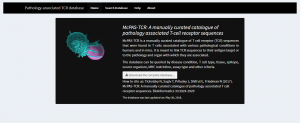
This database currently consists of more than 5000 pathologically-associated TCR sequences obtained from 118 published literature. A web-based search platform is user-friendly and allows users to search for the annotated TCR sequences using different criteria (https://rstudio.github.io/shinydashboard/index.html) Fig. 1. The complete database is also available for downloading.
Fig. 1 Homepage of McPAS-TCR web-based search tool [7].
The TCR sequences are classified into the following categories:
- TCRα and TCRβ chains
- Sequence category:
- Pathogens–bacteria, viruses, and parasites,
- Autoimmune–sequences identified in tissues/T-cells from human and mice with an autoimmune condition,
- Cancer–sequences identified in malignant tissues/T-cells of human origin, or in mice models of malignancies
- Allergy–sequences identified in allergic reactions to various allergens, and
- Other–sequences not classified to any of the above categories.
- Pathology
- Additional details
- Antigen identification method
- Next-generation sequencing (NGS)
- Antigen protein
- Epitope protein
- Major histocompatibility complex (MHC)
- Tissue
- Type of T-cell
- PubMed ID
This manually curated database can provide epitope specific TCR sequences easily and can be used for comparative analysis of the sequences recognizing the same epitope, disease-associated TCR sequences along with the links to the previous studies.
For further reading, click here.
References
- Davis, M. M., & Bjorkman, P. J. (1988). T-cell antigen receptor genes and T-cell recognition. Nature, 334(6181), 395.
- Bassing, C. H., Swat, W., & Alt, F. W. (2002). The mechanism and regulation of chromosomal V (D) J recombination. Cell, 109(2), S45-S55.
- Laydon, D. J., Bangham, C. R., & Asquith, B. (2015). Estimating T-cell repertoire diversity: limitations of classical estimators and a new approach. Phil. Trans. R. Soc. B, 370(1675), 20140291.
- Calis, J. J., & Rosenberg, B. R. (2014). Characterizing immune repertoires by high throughput sequencing: strategies and applications. Trends in immunology, 35(12), 581-590.
- Friedensohn, S., Khan, T. A., & Reddy, S. T. (2017). Advanced methodologies in high-throughput sequencing of immune repertoires. Trends in biotechnology, 35(3), 203-214.
- Heather, J. M., Ismail, M., Oakes, T., & Chain, B. (2017). High-throughput sequencing of the T-cell receptor repertoire: pitfalls and opportunities. Briefings in bioinformatics.
- Tickotsky, N., Sagiv, T., Prilusky, J., Shifrut, E., & Friedman, N. (2017). McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences. Bioinformatics, 33(18), 2924-2929.
Algorithms
miRBase: Explained
Micro RNAs (miRNAs) are the short endogenous RNAs (~22 nucleotides) and originate from the non-coding RNAs [1], produced in single-celled eukaryotes, viruses, plants, and animals [2]. miRNAs are capable of controlling homeostasis [2] and play significant roles in various biological processes such as degradation of mRNA and post-translational inhibition through complementary base pairing [3]. (more…)
Algorithms
Machine learning in prediction of ageing-related genes/proteins
Ageing has a great impact on human health, when people’s age advance towards 80 years, approximately half of the proteins in the body get damaged through oxidation. The chemical degradations occurring in our body produce energy by the consumed food via oxidation in the presence of oxygen. (more…)
Algorithms
Simulated sequence alignment software: An alternative to MSA benchmarks

In our previous article, we discussed different multiple sequence alignment (MSA) benchmarks to compare and assess the available MSA programs. However, since the last decade, several sequence simulation software have been introduced and are gaining more interest. In this article, we will be discussing various sequence simulating software being used as alternatives to MSA benchmarks. (more…)



You must be logged in to post a comment Login