Cheminformatics
How to do molecular orbital analysis to find d-orbitals involved in bonding in an organometallic compound?
Structure modeling of chemical compounds finds essential application in the field of cheminformatics. It is used to study the structural stability, metal-ion bonding, the presence of electrons, closed and open shell energies, the reactivity of complexes, molecular orbital analyzes, molecular mechanics, and so on. There is some software available for structural modeling of chemical compounds/complexes and the most widely used are Gaussian [1] and ORCA [2].
Gaussian software allows users to study molecular mechanics, ground-state semi-empirical calculations, Density functional theory (DFT), wave-function stability analysis (HT and DFT), electronic correlation, geometric optimization, vibrational frequency analysis, and so on.
In this tutorial, we will discuss Gaussian (G09) software and its application to perform molecular orbital analysis of a compound. This basic analysis requires an input file generated either using G09 or Avogadro. However, this tutorial will cover both the options and then finally the job will be run on G09 providing a detailed profile of orbitals in a compound/complex.
Let’s prepare the input file first.
1. Preparing the input file
The input file for the analysis can be generated by Gaussian itself but the coordinates need to be calculated and input manually, therefore, it’s easier to generate input file using Avogadro or ChemDraw and then open in Gaussian for further analysis. We will go through the steps for input file generation using both the software one by one.
i) using Avogadro
- Draw molecule.
- Go to ‘Extensions’ –> ‘Optimize Geometry’
- Go to ‘Extensions’ –> ‘Gaussian…’
- A window will appear as shown in Fig. 1, select the appropriate options as per your requirements. Our calculation is ‘Geometry Optimization’ and the other default parameters. After that, click ‘Generate’ and save the file as Gaussian Input Deck (.com) in the same folder.
Fig. 1 A screenshot showing the parameters of input file generation in Avogadro.
ii) using ChemDraw
- Draw molecule.
- Go to ‘Calculation’ –> ‘MM2’ (to optimize the geometry)
- Save the file as a .gjf file.
2. Running the job in Gaussian (G09)
- Open the G09.
- Go to ‘File’ –> ‘Open’. Open the .com/.gjf file generated using ChemDraw/ Avogadro.
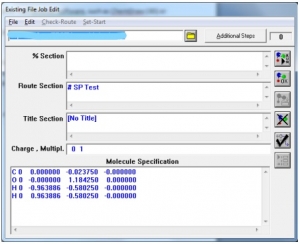
- A new window will appear showing different sections (Fig. 2).
Fig. 2 A screenshot of Gaussian displaying different sections for job submission.
The first line is the path of the file.
“%Section” specifies the name of checkpoint file.
“Route Section” is the main job defining section, where you can define which test you want to run. For this purpose, a specific keyword is used along with the command “Pop” explained as follows:
"Pop" = "Reg" # displays HOMO-5 up to LUMO+5 orbital information
"Pop" = "Full" # displays information of all orbitals
"Pop" = "NBO" # for Natural Bond Order analysis
"Pop" = "None" # no orbital information is displayed
"Pop" = "MK, CHELP, OR CHELPG" # produce charges fit to electrostatic potential (ESP)
“Title Section” allows the user to enter a job name.
“Charge, Multipl.” allows specifying the charge and spin multiplicity of the molecule separated by a space.
“Molecule specification” displays the atoms and their coordinates. These coordinates are calculated by Avogadro or ChemDraw else the user needs to calculate them manually.
That was about the input parameters, you can change them as per your requirements. Now, let’s get back to our job submission.
- Since we want molecular orbital analysis including the d-orbitals, therefore, we will use
Pop=Fullcommand. So, type the following in the Route Section:
Pop=Full FormCheck and click on ‘OK’ box present on the right side.
As you can see, another command FormCheck has been added. This command is used to create a file which contains the information of all molecular orbitals and can be plotted later.
- The memory is allocated suitably depending upon the configuration of your workstation.
- You can see the progress in the ‘Run Progress’ bar, it will be changed to ‘Ready to run processing start’.
- Now, to execute the g09 command, either click on
 or go to ‘Process’ –> ‘Begin Processing’.
or go to ‘Process’ –> ‘Begin Processing’. - After that, it will prompt for the folder where you want to save your output file. Click ‘Save’.
- Again, you will notice that the Run progress bar has changed the status to ‘C:\G09W\l1 is executing’.
This job takes a long time to finish, wait until it finishes providing the output files. These files can then be used to visualize the d-orbitals. The analyzes of output results will be explained in the upcoming articles.
References
- Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., … & Nakatsuji, H. (2009). Gaussian 09, revision A. 1. Gaussian Inc. Wallingford CT, 27, 34.
- Neese, F. (2012). The ORCA program system. Wiley Interdisciplinary Reviews: Computational Molecular Science, 2(1), 73-78.
Bioinformatics Programming
How to obtain ligand structures in PDB format from PDB ligand IDs?
Previously, we provided a similar script to download ligand SMILES from PDB ligand IDs. In this article, we are downloading PDB ligand structures from their corresponding IDs. (more…)
Cheminformatics
cheML.io: ML-generated molecules database

Due to the advancement of machine learning (ML) methods, we can find increasing applications of them in the field of bioinformatics as well. ML is being utilized in making personalized medicines, similarity searches in DNA and protein sequences, phylogenetics by mapping selected species on phylogenetic trees, gene and protein function annotation, generating chemical compounds, and so on. In this article, we will discuss an online database of ML-generated molecules known as cheML.io [1].
Bioinformatics Programming
How to obtain SMILES of ligands using PDB ligand IDs?
Fetching SMILE strings for a given number of SDF files of chemical compounds is not such a trivial task. We can quickly obtain them using RDKit or OpenBabel. But what if you don’t have SDF files of ligands in the first place? All you have is Ligand IDs from PDB. If they are a few then you can think of downloading SDF files manually but still, it seems time-consuming, especially when you have multiple compounds to work with. Therefore, we provide a Python script that will read all Ligand IDs and fetch their SDF files, and will finally convert them into SMILE strings. (more…)
Cheminformatics
Converting file formats using Openbabel.

Openbabel [1] offers a wide range of operations. One of which is file format conversion which is most widely used. In this article, we will describe commands that convert file formats. (more…)
Bioinformatics Programming
smitostr.py: Python script to convert SMILES to structures.
As mentioned in some of our previous articles, RDKit provides a wide range of functions. In this article, we are using RDKit [1] to draw a molecular structure using SMILES. (more…)
Bioinformatics Programming
tanimoto_similarities.py: A Python script to calculate Tanimoto similarities of multiple compounds using RDKit.
RDKit [1] is a very nice cheminformatics software. It allows us to perform a wide range of operations on chemical compounds/ ligands. We have provided a Python script to perform fingerprinting using Tanimoto similarity on multiple compounds using RDKit. (more…)
Cheminformatics
Installing CDK (Chemistry Development Kit) on Ubuntu (Linux)

CDK stands for chemistry development kit [1]. This is an open source kit for cheminformatics consisting of modular JAVA libraries. In this article, we will install CDK on Ubuntu. (more…)




You must be logged in to post a comment Login