

This is the second article under the popular series “Do you HYPHY…”, recently published online in BiR. In this issue, I would like to take the...
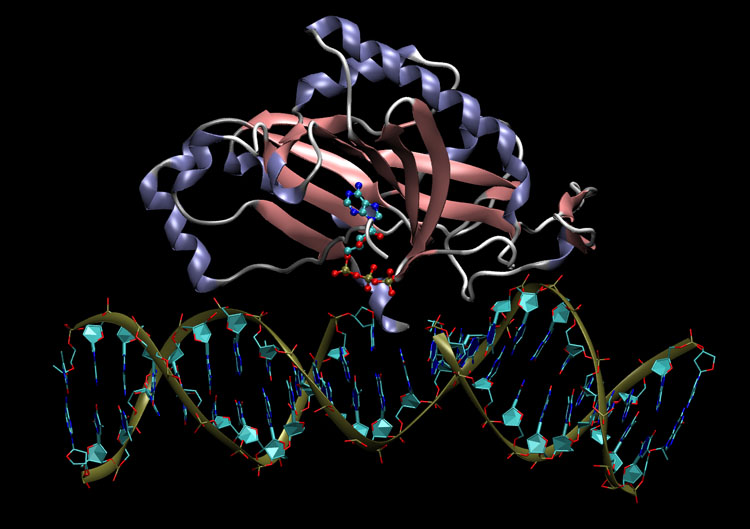

The identification and assignment of functions of unknown macro molecules has been observed to be faster and reliable than the sequence-based methods. This may be due...


As I have discussed in my earlier articles about the multiple sequence alignment (MSA) tools (MUSCLE & T-COFFEE). Now in this article, we will discuss different...


T-Coffee is a multiple sequence alignment tool which stands for Tree-based Consistency Objective Function for alignment Evaluation. It is a simultaneous alignment which combines the best...


HyPhy, acronym for Hypothesis Testing Using Phylogenies (www.hyphy.org) was written & designed by Kosakovsky Pond and workers to provide likelihood-based analyses on molecular evolutionary data sets...


Perl one-liners are extremely short Perl scripts written in the form of a string of commands that fits onto one line. That would amount to a...


The Microbial Pan Genome is the union of genes shared by genomes of interest. This term was first used by Medini in 2005.


As the needs say the importance of sequencing of genomes, it is equally important to visualize them. There exists some tools to visualize the genomes,but they...


In bioinformatics, GROMACS is one of the most popular Molecular Dynamics simulation software with loads of features built-in. Installing GROMACS Version 5.x.x+ can be a tedious...

After a lot of work in the field of bioinformatics, many of the living organisms’ genome has been sequenced and a lot of information has been...


CRISPR/Cas9 system is a bacterial defence mechanism against bacteriophage infection. When a viral dna(Bacteriophage, in this case) integrates into the bacterial genome, it produces RNA which...

In my last article I discussed about the Multiple Sequence Alignment and its creation. Now in this article, I am going to explain the workflow of...
Latest Comments